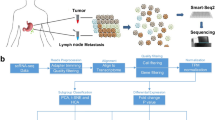
Abstract
Gastric carcinoma (GC) is regarded as one of the deadliest cancer characterized by diversity and haste metastasis and suffers limited understanding of the spatial variation between primary and metastatic GC tumors. In this project, transcriptome analysis on 46 primary tumorous, adjacent non-tumorous, and metastatic GC tissues was performed. The results demonstrated that metastatic tumorous tissues had diminished CD8+ T cells compared to primary tumors, which is mechanistically attributed to being due to innate immunity differences represented by marked differences in macrophages between metastatic and primary tumors, particularly those expressing ApoE, where their abundance is linked to unfavorable prognoses. Examining variations in gene expression and interactions indicated possible strategies of immune evasion hindering the growth of CD8+ T cells in metastatic tumor tissues. More insights could be gained into the immune evasion mechanisms by portraying information about the GC ecosystem.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, et al. Cancer incidence and mortality in China, 2016. J Natl Cancer Cent. 2022;2:1–9.
Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26–38.
Dolcetti R, De Re V, Canzonieri V. Immunotherapy for gastric cancer: time for a personalized approach? Int J Mol Sci. 2018;19:1602.
Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177:1330–1345.e18.
House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020;26:487–504.
Gao Y, Bado I, Wang H, Zhang W, Rosen JM, Zhang XH. Metastasis organotropism: redefining the congenial soil. Dev Cell. 2019;49:375–91.
Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308.e36.
Eruslanov E, Neuberger M, Daurkin I, Perrin GQ, Algood C, Dahm P, et al. Circulating and tumor-infiltrating myeloid cell subsets in patients with bladder cancer. Int J Cancer. 2012;130:1109–19.
Jiang H, Yu D, Yang P, Guo R, Kong M, Gao Y, et al. Revealing the transcriptional heterogeneity of organ-specific metastasis in human gastric cancer using single-cell RNA sequencing. Clin Transl Med. 2022;12:e730.
Sun Y, Wu L, Zhong Y, Zhou K, Hou Y, Wang Z, et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184:404–421.e16.
Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11:2285.
Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallée VP, et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020;26:259–69.
Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–72.
Bassez A, Vos H, Van Dyck L, Floris G, Arijs I, Desmedt C, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021;27:820–32.
Lu B, Ferrandino AF, Flavell RA. Gadd45beta is important for perpetuating cognate and inflammatory signals in T cells. Nat Immunol. 2004;5:38–44.
Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y, et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci USA. 2019;116:9999–10008.
van Hooren L, Handgraaf SM, Kloosterman DJ, Karimi E, van Mil L, Gassama AA, et al. CD103(+) regulatory T cells underlie resistance to radio-immunotherapy and impair CD8(+) T cell activation in glioblastoma. Nat Cancer. 2023;4:665–81.
Li X, Sun Z, Peng G, Xiao Y, Guo J, Wu B, et al. Single-cell RNA sequencing reveals a pro-invasive cancer-associated fibroblast subgroup associated with poor clinical outcomes in patients with gastric cancer. Theranostics. 2022;12:620–38.
Zhang L, Romero P. Metabolic control of CD8(+) T cell fate decisions and antitumor immunity. Trends Mol Med. 2018;24:30–48.
Stephen TL, Rutkowski MR, Allegrezza MJ, Perales-Puchalt A, Tesone AJ, Svoronos N, et al. Transforming growth factor β-mediated suppression of antitumor T cells requires FoxP1 transcription factor expression. Immunity 2014;41:427–39.
Wei H, Geng J, Shi B, Liu Z, Wang YH, Stevens AC, et al. Cutting edge: Foxp1 controls naive CD8+ T cell quiescence by simultaneously repressing key pathways in cellular metabolism and cell cycle progression. J Immunol. 2016;196:3537–41.
Papadakis KA, Krempski J, Reiter J, Svingen P, Xiong Y, Sarmento OF, et al. Krüppel-like factor KLF10 regulates transforming growth factor receptor II expression and TGF-β signaling in CD8+ T lymphocytes. Am J Physiol Cell Physiol. 2015;308:C362–71.
Liu P, Li P, Burke S. Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol Rev. 2010;238:138–49.
Rindler K, Jonak C, Alkon N, Thaler FM, Kurz H, Shaw LE, et al. Single-cell RNA sequencing reveals markers of disease progression in primary cutaneous T-cell lymphoma. Mol Cancer. 2021;20:124.
Hu B, Yu M, Ma X, Sun J, Liu C, Wang C, et al. IFNα potentiates Anti-PD-1 efficacy by remodeling glucose metabolism in the hepatocellular carcinoma microenvironment. Cancer Discov. 2022;12:1718–41.
Madden MZ, Rathmell JC. The complex integration of t-cell metabolism and immunotherapy. Cancer Discov. 2021;11:1636–43.
Amsen D, van Gisbergen K, Hombrink P, van Lier RAW. Tissue-resident memory T cells at the center of immunity to solid tumors. Nat Immunol. 2018;19:538–46.
Japanese Gastric Cancer A. Japanese gastric cancer treatment guidelines 2018 (5th edition). Gastric Cancer. 2021;24:1–21.
Xie C, Zhou X, Liang C, Li X, Ge M, Chen Y, et al. Apatinib triggers autophagic and apoptotic cell death via VEGFR2/STAT3/PD-L1 and ROS/Nrf2/p62 signaling in lung cancer. J Exp Clin Cancer Res. 2021;40:266.
Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988.e16.
Dos Santos TA, da Silva AC, Silva EB, Gomes PA, EspÃndola JW, Cardoso MV, et al. Antitumor and immunomodulatory activities of thiosemicarbazones and 1,3-Thiazoles in Jurkat and HT-29 cells. Biomed Pharmacother. 2016;82:555–60.
García-Mulero S, Alonso MH, Pardo J, Santos C, Sanjuan X, Salazar R, et al. Lung metastases share common immune features regardless of primary tumor origin. J Immunother Cancer. 2020;8:e000491.
Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 2022;12:134–53.
Karimi E, Yu MW, Maritan SM, Perus LJM, Rezanejad M, Sorin M, et al. Single-cell spatial immune landscapes of primary and metastatic brain tumours. Nature. 2023;614:555–63.
Kemp SB, Carpenter ES, Steele NG, Donahue KL, Nwosu ZC, Pacheco A, et al. Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-κB-mediated production of CXCL1. Cancer Res. 2021;81:4305–18.
Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502.
Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–82.
Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–6.
Funding
We express our gratitude to the patients for their support, as well as the National Natural Science Foundation of China (81802370, 82172925, 82072575) and the Youth Foundations of Zhongshan Hospital Fudan University (2022ZSQN09, 2023ZYYS-016) for their financial assistance.
Author information
Authors and Affiliations
Contributions
LTS and ZPF conceived the project, designed the research, and supervised the experiments; DY acquired data together with HKS; ZJY and LML analyzed and interpreted data; YYT, SX, XZH, LSY and ZYJ collected and assembled data; ZMX revised the manuscript for important intellectual content; LTS and ZPF obtained funding and provided technical and material support.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The Ethical Review Board of Zhongshan Hospital has reviewed this study, with approval NO. B2022-540R, and patients have provided informed consent.
Consent for publication
All authors approved the final manuscript and submission to the journal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dong, Y., Hu, K., Zhang, J. et al. ScRNA-seq of gastric cancer tissues reveals differences in the immune microenvironment of primary tumors and metastases. Oncogene (2024). https://doi.org/10.1038/s41388-024-03012-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41388-024-03012-5