Abstract
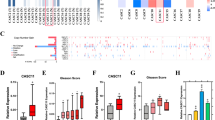
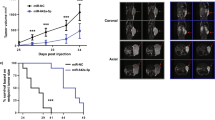
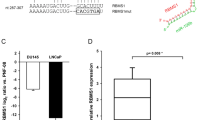
Prostate cancer (PCa) remains a significant cause of morbidity and mortality among men worldwide. A number of genes have been implicated in prostate tumorigenesis, but the mechanisms underlying their dysregulation are still incompletely understood. Evidence has established the competing endogenous RNA (ceRNA) theory as a novel regulatory mechanism for post-transcriptional alterations. Yet, a comprehensive characterization of ceRNA network in PCa lacks. Here we utilize stringent in-silico methods to construct a large ceRNA network across different PCa stages, and provide experimental demonstration for the competing regulation among protumorigenic SEC23A, PHTF2, and their corresponding ceRNA pairs. Using machine learning, we establish a ceRNA-based signature (ceRNA_sig) predictive of androgen receptor (AR) activity, tumor aggressiveness, and patient outcomes. Importantly, we identify miR-375 as a key node in PCa ceRNA network, which is upregulated in PCa relative to normal tissues. Forced expression of miR-375 significantly inhibits, while its inhibition promotes, aggressive behaviors of both AR+ and AR− PCa cells in vitro and in vivo. Mechanistically, we show that miR-375 predominantly targets genes possessing oncogenic roles (e.g., proliferation, DNA repair, and metastasis), and thus release targets with tumor suppressive functions. This action model well clarifies why an upregulated miRNA plays a tumor suppressive role in PCa. Together, our study provides new insights into understanding of transcriptomic aberrations during PCa evolution, and nominates miR-375 as a potential therapeutic target for combating aggressive PCa.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The RNA-seq data have been deposited in GEO database under accession code GSE234110.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Zhang D, Zhao S, Li X, Kirk JS, Tang DG. Prostate luminal progenitor cells in development and cancer. Trends Cancer. 2018;4:769–83.
Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701–11.
Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:10798.
Zhang D, Hu Q, Liu X, Ji Y, Chao HP, Liu Y, et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 2020;11:2089.
Zou C, He Q, Feng Y, Chen M, Zhang D. A m(6)Avalue predictive of prostate cancer stemness, tumor immune landscape and immunotherapy response. NAR Cancer. 2022;4:zcac010.
Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46.
Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54.
Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjostrom M, et al. The DNA methylation landscape of advanced prostate cancer. Nat. Genetics. 2020;52:778–89.
Li Q, Deng Q, Chao HP, Liu X, Lu Y, Lin K, et al. Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses. Nat Commun. 2018;9:3600.
Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Leopold V, et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci USA. 2013;110:7154–9.
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92.
Wei Y, Tang X, Ren Y, Yang Y, Song F, Fu J, et al. An RNA-RNA crosstalk network involving HMGB1 and RICTOR facilitates hepatocellular carcinoma tumorigenesis by promoting glutamine metabolism and impedes immunotherapy by PD-L1+ exosomes activity. Signal Transduct Targeted Ther. 2021;6:421.
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289.
Chan JJ, Kwok ZH, Chew XH, Zhang B, Liu C, Soong TW, et al. A FTH1 gene:pseudogene:microRNA network regulates tumorigenesis in prostate cancer. Nucleic Acids Res. 2018;46:1998–2011.
Yang G, Chen X, Quan Z, Liu M, Guo Y, Tang Y, et al. Comprehensive analysis of the FOXA1-related ceRNA network and identification of the MAGI2-AS3/DUSP2 axis as a prognostic biomarker in prostate cancer. Front Oncol. 2023;13:1048521.
Wang J, Huang J, Guo Y, Fu Y, Cao Y, Zhou K, et al. Identification and functional analysis of LncRNA-XIST ceRNA network in prostate cancer. BMC Cancer. 2022;22:935.
Wang Y, Lieberman R, Pan J, Zhang Q, Du M, Zhang P, et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol Cancer. 2016;15:70.
Selth LA, Das R, Townley SL, Coutinho I, Hanson AR, Centenera MM, et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene. 2017;36:24–34.
Gan J, Liu S, Zhang Y, He L, Bai L, Liao R, et al. MicroRNA-375 is a therapeutic target for castration-resistant prostate cancer through the PTPN4/STAT3 axis. Exp Mol Med. 2022;54:1290–305.
Cancer Genome Atlas Research N. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28.
Macedo-Silva C, Benedetti R, Ciardiello F, Cappabianca S, Jeronimo C, Altucci L. Epigenetic mechanisms underlying prostate cancer radioresistance. Clin Epigenetics. 2021;13:125.
Bushweller JH. Targeting transcription factors in cancer - from undruggable to reality. Nat Rev Cancer. 2019;19:611–24.
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386.
Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–36.
Thomas-Jardin SE, Dahl H, Nawas AF, Bautista M, Delk NA. NF-kappaB signaling promotes castration-resistant prostate cancer initiation and progression. Pharmacol Ther. 2020;211:107538.
Chan JM, Zaidi S, Love JR, Zhao JL, Setty M, Wadosky KM, et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science. 2022;377:1180–91.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Xiong J, Yan L, Zou C, Wang K, Chen M, Xu B, et al. Integrins regulate stemness in solid tumor: an emerging therapeutic target. J Hematol Oncol. 2021;14:177.
Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305.
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–62.
Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71:5950–4.
Ouyang Y, Gao P, Zhu B, Chen X, Lin F, Wang X, et al. Downregulation of microRNA-429 inhibits cell proliferation by targeting p27Kip1 in human prostate cancer cells. Mol Med Rep. 2015;11:1435–41.
Sharma S, Pei X, Xing F, Wu SY, Wu K, Tyagi A, et al. Regucalcin promotes dormancy of prostate cancer. Oncogene. 2021;40:1012–26.
Bhagirath D, Liston M, Patel N, Akoto T, Lui B, Yang TL, et al. MicroRNA determinants of neuroendocrine differentiation in metastatic castration-resistant prostate cancer. Oncogene. 2020;39:7209–23.
Fu Y, Cao T, Zou X, Ye Y, Liu Y, Peng Y, et al. AKT1 regulates UHRF1 protein stability and promotes the resistance to abiraterone in prostate cancer. Oncogenesis. 2023;12:1.
Zeng B, Zhao Q, Sun Z, Liu D, Chen H, Li X, et al. SEC23A is an independent prognostic biomarker in bladder cancer correlated With MAPK signaling. Front Genet. 2021;12:672832.
Chi Y, Wang H, Wang F, Ding M. PHTF2 regulates lipids metabolism in gastric cancer. Aging. 2020;12:6600–10.
Lu X, Fong KW, Gritsina G, Wang F, Baca SC, Brea LT, et al. HOXB13 suppresses de novo lipogenesis through HDAC3-mediated epigenetic reprogramming in prostate cancer. Nat Genetics. 2022;54:670–83.
Jing J, Wang B, Liu P. The functional role of SEC23 in vesicle transportation, autophagy and cancer. Int J Biol Sci. 2019;15:2419–26.
Cheng Q, Liu K, Xiao J, Shen K, Wang Y, Zhou X, et al. SEC23A confers ER stress resistance in gastric cancer by forming the ER stress-SEC23A-autophagy negative feedback loop. J Exp Clin Cancer Res. 2023;42:232.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162:454.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126.
Wu W, Li Q, Zhu Z, Li C, Lu P, Zhou X, et al. HTR1D functions as a key target of HOXA10-AS/miR-340-3p axis to promote the malignant outcome of pancreatic cancer via PI3K-AKT signaling pathway. Int J Biol Sci. 2022;18:3777–94.
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612–26.
Yao B, Zhu S, Wei X, Chen MK, Feng Y, Li Z, et al. The circSPON2/miR-331-3p axis regulates PRMT5, an epigenetic regulator of CAMK2N1 transcription and prostate cancer progression. Mol Cancer. 2022;21:119.
Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47.
Wu Y, Sun X, Song B, Qiu X, Zhao J. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. 2017;6:1686–97.
He Z, Li W, Zheng T, Liu D, Zhao S. Human umbilical cord mesenchymal stem cells-derived exosomes deliver microRNA-375 to downregulate ENAH and thus retard esophageal squamous cell carcinoma progression. J Exp Clin Cancer Res. 2020;39:140.
Xu X, Liu Y, Li Y, Chen H, Zhang Y, Liu J, et al. Selective exosome exclusion of miR-375 by glioma cells promotes glioma progression by activating the CTGF-EGFR pathway. J Exp Clin Cancer Res. 2021;40:16.
Li Q, Liu B, Chao HP, Ji Y, Lu Y, Mehmood R, et al. LRIG1 is a pleiotropic androgen receptor-regulated feedback tumor suppressor in prostate cancer. Nat Commun. 2019;10:5494.
Braun DA, Hou Y, Bakouny Z, Ficial M, Sant’ Angelo M, Forman J, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med. 2020;26:909–18.
Lauss M, Donia M, Harbst K, Andersen R, Mitra S, Rosengren F, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat Commun. 2017;8:1738.
Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–49.e916.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7.
Huang HY, Lin YC, Cui S, Huang Y, Tang Y, Xu J, et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022;50:D222–D230.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97.
Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–152.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005.
Funding
This work was supported by grants from the Excellent Youth Foundation of Hunan Province (2021JJ10028), the National Natural Science Foundation of China (81972418), the Shenzhen Natural Science Foundation (JCYJ20220530160410024), and the Fundamental Research Funds for the Central Universities. CZ was supported, in part, by the Hunan Province Natural Science Foundation (2022JJ40111) and the Changsha Natural Science Foundation (kq2202182).
Author information
Authors and Affiliations
Contributions
CZ and DZ conceived and designed the study, analyzed, and interpreted data, and finalized the manuscript. MC and YT performed most experiments with help from YL Specifically, WL provided the clinical samples and performed IHC analysis. CZ conducted Bioinformatic analysis with input from MC MC and CZ initially drafted the manuscript. All authors read and approved the manuscript. We apologize to the colleagues whose work was not cited due to space constraint.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, M., Zou, C., Tian, Y. et al. An integrated ceRNA network identifies miR-375 as an upregulated miRNA playing a tumor suppressive role in aggressive prostate cancer. Oncogene (2024). https://doi.org/10.1038/s41388-024-03011-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41388-024-03011-6