Abstract
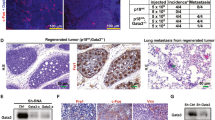
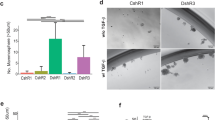
Glycoprotein Nmb (GPNMB) is overexpressed in triple-negative and basal-like breast cancers and its expression is predictive of poor prognosis within this aggressive breast cancer subtype. GPNMB promotes breast cancer growth, invasion, and metastasis; however, its role in mammary tumor initiation remains unknown. To address this question, we overexpressed GPNMB in the mammary epithelium to generate MMTV/GPNMB transgenic mice and crossed these animals to the MMTV/Wnt-1 mouse model, which is known to recapitulate features of human basal breast cancers. We show that GPNMB alone does not display oncogenic properties; however, its expression dramatically accelerates tumor onset in MMTV/Wnt-1 mice. MMTV/Wnt-1 × MMTV/GPNMB bigenic mice also exhibit a significant increase in the growth rate of established primary tumors, which is attributable to increased proliferation and decreased apoptosis. To elucidate molecular mechanisms underpinning the tumor-promoting effects of GPNMB in this context, we interrogated activated pathways in tumors derived from the MMTV/Wnt-1 and MMTV/Wnt-1 × MMTV/GPNMB mice using RPPA analysis. These data revealed that MMTV/Wnt-1 × MMTV/GPNMB bigenic tumors exhibit a pro-growth signature characterized by elevated PI3K/AKT/mTOR signaling and increased β-catenin activity. Furthermore, we extended these observations to an independent Wnt-1 expressing model of aggressive breast cancer, and confirmed that GPNMB enhances canonical Wnt pathway activation, as evidenced by increased β-catenin transcriptional activity, in breast cancer cells and tumors co-expressing Wnt-1 and GPNMB. GPNMB-dependent engagement of β-catenin occurred, in part, through AKT activation. Taken together, these data ascribe a novel, pro-growth role for GPNMB in Wnt-1 expressing basal breast cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–34.
Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–33.
Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23.
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90.
Sharma P. Biology and Management of Patients With Triple-Negative Breast Cancer. Oncologist. 2016;21:1050–62.
Maric G, Rose AA, Annis MG, Siegel PM. Glycoprotein non-metastatic b (GPNMB): A metastatic mediator and emerging therapeutic target in cancer. Onco Targets Ther. 2013;6:839–52.
Rose AAN, Biondini M, Curiel R, Siegel PM. Targeting GPNMB with glembatumumab vedotin: Current developments and future opportunities for the treatment of cancer. Pharmacol Ther. 2017;179:127–41.
Rose AA, Grosset AA, Dong Z, Russo C, Macdonald PA, Bertos NR, et al. Glycoprotein nonmetastatic B is an independent prognostic indicator of recurrence and a novel therapeutic target in breast cancer. Clin Cancer Res: Off J Am Assoc Cancer Res. 2010;16:2147–56.
Rose AA, Annis MG, Dong Z, Pepin F, Hallett M, Park M, et al. ADAM10 releases a soluble form of the GPNMB/Osteoactivin extracellular domain with angiogenic properties. PLoS ONE. 2010;5:e12093.
Kanematsu M, Futamura M, Takata M, Gaowa S, Yamada A, Morimitsu K, et al. Clinical significance of glycoprotein nonmetastatic B and its association with HER2 in breast cancer. Cancer Med. 2015;4:1344–55.
Maric G, Annis MG, Dong Z, Rose AA, Ng S, Perkins D, et al. GPNMB cooperates with neuropilin-1 to promote mammary tumor growth and engages integrin ɑ5β1 for efficient breast cancer metastasis. Oncogene. 2015;34:5494–5504.
Rose AA, Pepin F, Russo C, Abou Khalil JE, Hallett M, Siegel PM. Osteoactivin promotes breast cancer metastasis to bone. Mol Cancer Res. 2007;5:1001–14.
Okita Y, Kimura M, Xie R, Chen C, Shen LT, Kojima Y, et al. The transcription factor MAFK induces EMT and malignant progression of triple-negative breast cancer cells through its target GPNMB. Sci Signal. 2017;10:474.
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–24.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76.
Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013;14:R125.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74.
Li Y, Hively WP, Varmus HE. Use of MMTV-Wnt-1 transgenic mice for studying the genetic basis of breast cancer. Oncogene. 2000;19:1002–9.
Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100:15853–8.
Huang S, Li Y, Chen Y, Podsypanina K, Chamorro M, Olshen AB, et al. Changes in gene expression during the development of mammary tumors in MMTV-Wnt-1 transgenic mice. Genome Biol. 2005;6:R84.
Howe LR, Brown AM. Wnt signaling and breast cancer. Cancer Biol Ther. 2004;3:36–41.
van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50.
Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176:2911–20.
Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, et al. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24:209–31.
Lopez-Knowles E, O’Toole SA, McNeil CM, Millar EK, Qiu MR, Crea P, et al. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer. 2010;126:1121–31.
Li S, Li S, Sun Y, Li L. The expression of beta-catenin in different subtypes of breast cancer and its clinical significance. Tumour Biol. 2014;35:7693–8.
Bankfalvi A, Ludwig A, De-Hesselle B, Buerger H, Buchwalow IB, Boecker W. Different proliferative activity of the glandular and myoepithelial lineages in benign proliferative and early malignant breast diseases. Mod Pathol. 2004;17:1051–61.
Bocker W, Moll R, Poremba C, Holland R, Van Diest PJ, Dervan P, et al. Common adult stem cells in the human breast give rise to glandular and myoepithelial cell lineages: a new cell biological concept. Lab Invest. 2002;82:737–46.
Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7:659–72.
Nieto AI, Shyamala G, Galvez JJ, Thordarson G, Wakefield LM, Cardiff RD. Persistent mammary hyperplasia in FVB/N mice. Comp Med. 2003;53:433–8.
Wakefield LM, Thordarson G, Nieto AI, Shyamala G, Galvez JJ, Anver MR, et al. Spontaneous pituitary abnormalities and mammary hyperplasia in FVB/NCr mice: implications for mouse modeling. Comp Med. 2003;53:424–32.
Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21.
Robles AI, Varticovski L. Harnessing genetically engineered mouse models for preclinical testing. Chem Biol Interact. 2008;171:159–64.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7.
Huang S, Chen Y, Podsypanina K, Li Y. Comparison of expression profiles of metastatic versus primary mammary tumors in MMTV-Wnt-1 and MMTV-Neu transgenic mice. Neoplasia. 2008;10:118–24.
Maubant S, Tesson B, Maire V, Ye M, Rigaill G, Gentien D, et al. Transcriptome analysis of Wnt3a-treated triple-negative breast cancer cells. PLoS ONE. 2015;10:e0122333.
Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74.
Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22.
Failor KL, Desyatnikov Y, Finger LA, Firestone GL. Glucocorticoid-induced degradation of glycogen synthase kinase-3 protein is triggered by serum- and glucocorticoid-induced protein kinase and Akt signaling and controls beta-catenin dynamics and tight junction formation in mammary epithelial tumor cells. Mol Endocrinol. 2007;21:2403–15.
Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–99.
Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–48.
Xu J, Chen Y, Huo D, Khramtsov A, Khramtsova G, Zhang C, et al. beta-catenin regulates c-Myc and CDKN1A expression in breast cancer cells. Mol Carcinog. 2016;55:431–9.
Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4:11.
Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–57.
Watanabe K, Biesinger J, Salmans ML, Roberts BS, Arthur WT, Cleary M, et al. Integrative ChIP-seq/microarray analysis identifies a CTNNB1 target signature enriched in intestinal stem cells and colon cancer. PLoS ONE. 2014;9:e92317.
Chen C, Okita Y, Watanabe Y, Abe F, Fikry MA, Ichikawa Y, et al. Glycoprotein nmb Is Exposed on the Surface of Dormant Breast Cancer Cells and Induces Stem Cell-like Properties. Cancer Res. 2018;78:6424–35.
Ding VW, Chen RH, McCormick F. Differential regulation of glycogen synthase kinase 3beta by insulin and Wnt signaling. J Biol Chem. 2000;275:32475–81.
Perry JM, He XC, Sugimura R, Grindley JC, Haug JS, Ding S, et al. Cooperation between both Wnt/{beta}-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev. 2011;25:1928–42.
Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;7:e1000121.
Mulholland DJ, Dedhar S, Wu H, Nelson CC. PTEN and GSK3beta: key regulators of progression to androgen-independent prostate cancer. Oncogene. 2006;25:329–37.
Yuan H, Mao J, Li L, Wu D. Suppression of glycogen synthase kinase activity is not sufficient for leukemia enhancer factor-1 activation. J Biol Chem. 1999;274:30419–23.
Davies BR, Greenwood H, Dudley P, Crafter C, Yu DH, Zhang J, et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol Cancer Ther. 2012;11:873–87.
Lin J, Sampath D, Nannini MA, Lee BB, Degtyarev M, Oeh J, et al. Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin Cancer Res: Off J Am Assoc Cancer Res. 2013;19:1760–72.
Donehower LA, Godley LA, Aldaz CM, Pyle R, Shi YP, Pinkel D, et al. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–95.
Kapoun AM, Shackleford GM. Preferential activation of Fgf8 by proviral insertion in mammary tumors of Wnt1 transgenic mice. Oncogene. 1997;14:2985–9.
Kwan H, Pecenka V, Tsukamoto A, Parslow TG, Guzman R, Lin TP, et al. Transgenes expressing the Wnt-1 and int-2 proto-oncogenes cooperate during mammary carcinogenesis in doubly transgenic mice. Mol Cell Biol. 1992;12:147–54.
Li Y, Podsypanina K, Liu X, Crane A, Tan LK, Parsons R, et al. Deficiency of Pten accelerates mammary oncogenesis in MMTV-Wnt-1 transgenic mice. BMC Mol Biol. 2001;2:2.
Podsypanina K, Li Y, Varmus HE. Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype. BMC Med. 2004;2:24.
Shackleford GM, MacArthur CA, Kwan HC, Varmus HE. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci USA. 1993;90:740–4.
Jin R, Jin YY, Tang YL, Yang HJ, Zhou XQ, Lei Z. GPNMB silencing suppresses the proliferation and metastasis of osteosarcoma cells by blocking the PI3K/Akt/mTOR signaling pathway. Oncol Rep. 2018;39:3034–40.
Ono Y, Chiba S, Yano H, Nakayama N, Saio M, Tsuruma K, et al. Glycoprotein nonmetastatic melanoma protein B (GPNMB) promotes the progression of brain glioblastoma via Na(+)/K(+)-ATPase. Biochem Biophys Res Commun. 2016;481:7–12.
Yu B, Sondag GR, Malcuit C, Kim MH, Safadi FF. Macrophage-Associated Osteoactivin/GPNMB Mediates Mesenchymal Stem Cell Survival, Proliferation, and Migration Via a CD44-Dependent Mechanism. J Cell Biochem. 2016;117:1511–21.
Nagahara Y, Shimazawa M, Ohuchi K, Ito J, Takahashi H, Tsuruma K, et al. GPNMB ameliorates mutant TDP-43-induced motor neuron cell death. J Neurosci Res. 2017;95:1647–65.
Bao G, Wang N, Li R, Xu G, Liu P, He B. Glycoprotein non-metastaticmelanoma protein B promotes glioma motility and angiogenesis through the Wnt/beta-catenin signaling pathway. Exp Biol Med (Maywood). 2016;241:1968–76.
Zhang YX, Qin CP, Zhang XQ, Wang QR, Zhao CB, Yuan YQ, et al. Knocking down glycoprotein nonmetastatic melanoma protein B suppresses the proliferation, migration, and invasion in bladder cancer cells. Tumour Biol. 2017;39:1010428317699119.
Musgrove EA. Wnt signalling via the epidermal growth factor receptor: a role in breast cancer? Breast Cancer Res. 2004;6:65–8.
Hu T, Li C. Convergence between Wnt-beta-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236.
Schroeder JA, Troyer KL, Lee DC. Cooperative induction of mammary tumorigenesis by TGFalpha and Wnts. Oncogene. 2000;19:3193–9.
Schroeder JA, Adriance MC, McConnell EJ, Thompson MC, Pockaj B, Gendler SJ. ErbB-beta-catenin complexes are associated with human infiltrating ductal breast and murine mammary tumor virus (MMTV)-Wnt-1 and MMTV-c-Neu transgenic carcinomas. J Biol Chem. 2002;277:22692–8.
Yardley DA, Weaver R, Melisko ME, Saleh MN, Arena FP, Forero A, et al. EMERGE: A randomized phase II study of the antibody-drug conjugate glembatumumab vedotin in advanced glycoprotein NMB-expressing breast cancer. J Clin Oncol. 2015;33:1609–19.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Iadevaia S, Lu Y, Morales FC, Mills GB, Ram PT. Identification of optimal drug combinations targeting cellular networks: integrating phospho-proteomics and computational network analysis. Cancer Res. 2010;70:6704–14.
Northey JJ, Chmielecki J, Ngan E, Russo C, Annis MG, Muller WJ, et al. Signaling through ShcA is required for transforming growth factor beta- and Neu/ErbB-2-induced breast cancer cell motility and invasion. Mol Cell Biol. 2008;28:3162–76.
Acknowledgements
We acknowledge the Goodman Cancer Research Centre (GCRC) transgenic facility (McGill University) for generation of the MMTV/GPNMB mice, the GCRC histology core facility (McGill University) for routine histological services and Zhifeng Dong for some Immunohistochemical staining. We thank members of the Siegel laboratory for thoughtful discussions and critical reading of the manuscript. GM acknowledges studentship support from the Canadian Institutes for Health Research (CIHR). PMS is currently a McGill University William Dawson Scholar. This research was supported solely by grants from the CIHR (MOP-119401; PJT-153327).
Author contributions
GM, MGA, and PMS designed research; GM and MGA performed experiments; PM, CR, MGA, and DP generated reagents used in this study; DRS and GM performed and analyzed the RPPA analysis; GM, MGA, and PMS analyzed/interpreted data and GM, MGA, and PMS wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41388_2019_793_MOESM2_ESM.pptx
Supplementary Figure 1. MMTV/GPNMB transgenic mice express GPNMB in the mammary ductal epithelium and display normal virgin mammary gland development
41388_2019_793_MOESM5_ESM.pptx
Supplementary Figure 4. Immunohistochemistry of MMTV/Wnt-1 and MMTV/Wnt-1 x MMTV/GPNMB tumors to validate targets of the mTOR pathway
41388_2019_793_MOESM6_ESM.pptx
Supplementary Figure 5. Immunoblot analysis of epithelial and mesenchymal markers in 4T1-533LM and transgenic derived mammary tumors
Rights and permissions
About this article
Cite this article
Maric, G., Annis, M.G., MacDonald, P.A. et al. GPNMB augments Wnt-1 mediated breast tumor initiation and growth by enhancing PI3K/AKT/mTOR pathway signaling and β-catenin activity. Oncogene 38, 5294–5307 (2019). https://doi.org/10.1038/s41388-019-0793-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0793-7
This article is cited by
-
CCN3/NOV promotes metastasis and tumor progression via GPNMB-induced EGFR activation in triple-negative breast cancer
Cell Death & Disease (2023)
-
Discovering the key genes and important DNA methylation regions in breast cancer
Hereditas (2022)
-
Macrophages and cancer stem cells: a malevolent alliance
Molecular Medicine (2021)
-
The soluble glycoprotein NMB (GPNMB) produced by macrophages induces cancer stemness and metastasis via CD44 and IL-33
Cellular & Molecular Immunology (2021)
-
Glembatumumab vedotin for patients with metastatic, gpNMB overexpressing, triple-negative breast cancer (“METRIC”): a randomized multicenter study
npj Breast Cancer (2021)