Abstract
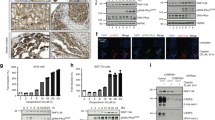
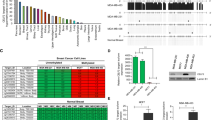
The point mutation that substitutes lysine with arginine at position 120 of human p53 has been characterized as a missense mutation. The K120R mutation renders the p53 protein disabled for acetylation and, as a result, defective for apoptotic function, which provides a mechanistic link between the missense mutation and tumorigenesis. However, we noticed the failures of tumorigenesis in mice with the mutation, and of the related studies to notice that it has arbitrarily reflected in amino acid change through a sequence modification (AGA) of the original tumor mutation (AGG) by codon degeneracy. Unlike this modified version, we also discovered a novel splicing site the original mutation, TP53 c.359A>G, may induce. Using a human induced pluripotent stem cell line that was engineered to be homozygous for the original mutation, we here identified that the accidental splicing site generates a defective transcript variant with a frame-shifted premature termination codon which is subjected to nonsense-mediated mRNA decay. The authentic splicing still occurs but in extremely low amounts. Taken together, this mutation causes depletion of cellular p53 via defective mRNA, suggesting a new link to tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Deissler H, Kafka A, Schuster E, Sauer G, Kreienberg R, Zeillinger R. Spectrum of p53 mutations in biopsies from breast cancer patients selected for preoperative chemotherapy analysed by the functional yeast assay to predict therapeutic response. Oncol Rep. 2004;11:1281–6.
Hashimoto T, Tokuchi Y, Hayashi M, Kobayashi Y, Nishida K, Hayashi S, et al. p53 null mutations undetected by immunohistochemical staining predict a poor outcome with early-stage non-small cell lung carcinomas. Cancer Res. 1999;59:5572–7.
Hayes VM, Dirven CM, Dam A, Verlind E, Molenaar WM, Mooij JJ, et al. High frequency of TP53 mutations in juvenile pilocytic astrocytomas indicates role of TP53 in the development of these tumors. Brain Pathol. 1999;9:463–7.
Jones S, Zhang XS, Parsons DW, Lin JCH, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6.
Leitao MM, Soslow RA, Baergen RN, Olvera N, Arroyo C, Boyd J. Mutation and expression of the TP53 gene in early stage epithelial ovarian carcinoma. Gynecol Oncol. 2004;93:301–6.
Meyers FJ, Chi SG, Fishman JR, deVere White RW, Gumerlock PH. p53 mutations in benign prostatic hyperplasia. J Natl Cancer Inst. 1993;85:1856–8.
Liu N, Wang J, Wang J, Wang R, Liu Z, Yu Y, et al. ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 2013;73:3749–60.
Monteith JA, Mellert H, Sammons MA, Kuswanto LA, Sykes SM, Resnick-Silverman L, et al. A rare DNA contact mutation in cancer confers p53 gain-of-function and tumor cell survival via TNFAIP8 induction. Mol Oncol. 2016;10:1207–20.
Shinmen N, Koshida T, Kumazawa T, Sato K, Shimada H, Matsutani T, et al. Activation of NFAT signal by p53-K120R mutant. FEBS Lett. 2009;583:1916–22.
Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–51.
Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–39.
Yun T, Yu K, Yang S, Cui Y, Wang Z, Ren H, et al. Acetylation of p53 protein at lysine 120 up-regulates Apaf-1 protein and sensitizes the mitochondrial apoptotic pathway. J Biol Chem. 2016;291:7386–95.
Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–83.
Liu XF, Tan YQ, Zhang CF, Zhang Y, Zhang LL, Ren PW, et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17:349–66.
Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–26.
Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–9.
Kim BY, Jeong S, Lee SY, Lee SM, Gweon EJ, Ahn H, et al. Concurrent progress of reprogramming and gene correction to overcome therapeutic limitation of mutant ALK2-iPSC. Exp Mol Med. 2016;48:e237.
Lee SY, Chung SK. Integrating gene correction in the reprogramming and transdifferentiation processes: a one-step strategy to overcome stem cell-based gene therapy limitations. Stem Cells Int. 2016;2016:2725670.
Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22.
Liebermann DA, Hoffman B, Vesely D. p53 induced growth arrest versus apoptosis and its modulation by survival cytokines. Cell Cycle. 2007;6:166–70.
Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99.
Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–50.
Gong L, Pan X, Lim CB, de Polo A, Little JB, Yuan ZM. A functional interplay between Delta 133p53 and Delta Np63 in promoting glycolytic metabolism to fuel cancer cell proliferation. Oncogene. 2018;37:2150–64.
Jayasinghe RG, Cao S, Gao Q, Wendl MC, Vo NS, Reynolds SM, et al. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 2018;23:270–81 e3.
Yu H, Koo I, Jeong S. Relative quantitation of restriction fragment length polymorphic DNAs via DNA melting analysis provides an effective way to determine allele frequencies. Genomics. 2009;94:355–61.
Chung SK, Lee MG, Ryu BK, Lee JH, Han J, Byun DS, et al. Frequent alteration of XAF1 in human colorectal cancers: implication for tumor cell resistance to apoptotic stresses. Gastroenterology. 2007;132:2459–77.
Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Tang J, Hu M, Lee S, Roblin R. A polymerase chain reaction based method for detecting mycoplasma/acholeplasma contaminants in cell culture. J Microbiol Methods. 2000;39:121–6.
Acknowledgements
We thank B-J. Park, K-S. Chae, and I. Horikawa for comments on the manuscript and Y-D. Kim for technical assistance. This research was supported in part by the National Research Foundation of Korea Grants (NRF-2013M3A9B4076487) and Korea Institute of Oriental Medicine Grants (K18131).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lee, SY., Park, JH., Jeong, S. et al. K120R mutation inactivates p53 by creating an aberrant splice site leading to nonsense-mediated mRNA decay. Oncogene 38, 1597–1610 (2019). https://doi.org/10.1038/s41388-018-0542-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0542-3
This article is cited by
-
Regulating tumor suppressor genes: post-translational modifications
Signal Transduction and Targeted Therapy (2020)