Abstract
Influenza viruses are highly transmissible, both within and between host species. The severity of the disease they cause is highly variable, from the mild and inapparent through to the devastating and fatal. The unpredictability of epidemic and pandemic outbreaks is accompanied but the predictability of seasonal disease in wide areas of the Globe, providing an inexorable toll on human health and survival. Although there have been great improvements in understanding influenza viruses and the disease that they cause, our knowledge of the effects they have on the host and the ways that the host immune system responds continues to develop. This review highlights the importance of the mucosa in defence against infection and in understanding the pathogenesis of disease. Although vaccines have been available for many decades, they remain suboptimal in needing constant redesign and in only providing short-term protection. There are real prospects for improvement in treatment and prevention of influenza soon, based on deeper knowledge of how the virus transmits, replicates and triggers immune defences at the mucosal surface.
Similar content being viewed by others
Introduction
In 1918, the Austrian artist Egon Schiele (Fig. 1) was finally achieving fame. He was beginning to take on the mantle of his better-known friend and mentor Gustav Klimt, who had died in February of that year. After years of struggling for recognition Schiele was at last able to afford a larger studio and an apartment in which to accommodate his wife and budding family. He was 28 years old.
High-res available from: https://www.egon-schiele.com/the-family.jsp.
By September 1918, so many Viennese key workers were sick with influenza that the military were called in to assist. By October, the pandemic had become so severe that Vienna’s schools were shut. Soon thereafter Egon Schiele’s wife, Edith, fell ill; six months pregnant, she and her unborn child died on the morning of October 28 after an illness lasting nine days. Three days later, Egon Schiele himself died of flu. A whole family, extinguished within a week.
The 1918-19 pandemic of H1N1 influenza not only affected schools and public services. It decimated the German and allied troops, infecting an estimated one-fifth of the global human population including the British Prime Minister David Lloyd George, US President Wilson, the French Prime Minister and the German Chancellor. In less than a year it killed about 50 million people, four times the number who died in the Great War.
Over one hundred years on and despite a vast amount of research,1 it is remarkable that knowledge of the precise pathogenesis of disease, optimal vaccines and appropriate specific treatments remain elusive. There is no doubt that the use of oxygen, mechanical ventilation, antivirals, antibiotics and the other modern medical armamentaria would have saved many of those who died from the 1918–1919 Spanish flu, but seasonal flu continues to kill vast numbers of people every year. At the centenary of the end of that great pandemic, it is timely to review what we now know and don’t know about both pandemic and seasonal influenza (see Table 1).
Influenza virus replication and key immune responses to infection
We will not discuss the virology of influenza replication (which is well described elsewhere, for example2) but in outline, after seeding in the respiratory mucosa, most influenza virus replication occurs in airway and alveolar epithelial cells. On entering the respiratory tract, the haemagglutinin (HA) proteins expressed on the surface of the influenza A or influenza B virus attach to sialic acid cell-surface receptors on the respiratory epithelium, triggering receptor-mediated endocytosis of the virion. This is followed by fusion of the virus membrane and the endosomal membrane, facilitated by HA and an acidic environment in the endosome. Ribonucleoprotein (RNP; containing the viral genome, across 8 viral RNA gene segments and the RNA-dependent RNA polymerase) is released and transported into the nucleus of the infected cell, where the RNA-dependent RNA polymerase mediates transcription and replication of viral RNA. The influenza virion consists of the RNP, is lined by the matrix protein 1 (M1) and is coated with the surface proteins HA, neuraminidase (NA) and matrix protein 2 (M2) that have been manufactured by the infected cell and then expressed on the cell surface.3 Once this packaging and budding process is complete, new virions are cleaved from the sialic acid residues on the cell surface by NA on progeny virions; the infected cell dies and the released virions can proceed to infect other cells.
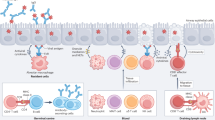
The sequence of events in the host is summarised in Fig. 2. As soon as influenza virus encounters host cells, an innate immune response is triggered. This is facilitated by pattern recognition receptors such as retinoic acid-inducible gene-I protein and toll-like receptors.4 The production and release of soluble immune mediators that ensues is typically comprised of antiviral responses that involve pro-inflammatory cytokines and interferons, and the transcription of many interferon-stimulated genes, the products of which target and interrupt multiple, different steps in the influenza virus replication cycle.5 At this stage infection may be prevented by such innate immune responses, or become established, leading to viral replication. Infection may also be prevented by pre-existing antibodies, particularly those at the respiratory site of viral entry such as IgA.
Viral entry and infection of the respiratory epithelium is blocked by specific mucosal antibody, mucus and antimicrobial proteins. Once access is gained to mucosal cells, inflammatory mediators produced. This initial phase is influenced by genetic factors, environmental stimuli, the resident respiratory microbiome, and infection history. Innate responses by resident airway cells, macrophages, and NK cells impede viral replication and spread to other parts of the respiratory tract. T-cell responses are important for viral clearance and disease resolution but may be associated with inappropriately polarized responses and immunopathology.
Infected respiratory epithelial cells activate the recruitment of NK cells, neutrophils and monocytes, which interact directly with infected cells to limit virus replication and sometimes act indirectly by priming adaptive immune cells and initiating the migration of other innate immune cells including dendritic cells, macrophages and natural killer cells.6 Collectively, these innate immune responses help to control and eliminate the influenza virus infection, but can also contribute to the development of symptoms and immunopathology.7
As viral replication comes under control, the adaptive immune response takes a more prominent role. Cytotoxic T lymphocytes both produce interferon gamma and target virus-infected cells for destruction.8 CD4+ T cells also target infected cells, facilitating subsequent activation of B cells and the production of anti-influenza antibodies.9 In addition, CD4+ T cells differentiate into Th1, Th2, Th17, follicular helper T cells and regulatory T cells; together, these differentiated cells have specific roles in antiviral soluble immune mediator response, activation of innate effector cells, promotion of B cell responses and regulation of cellular immune responses during influenza virus infection.9,10
The adaptive response exhibits memory, preventing future infections with the same strain of influenza. During re-exposure, T and B cells responses are faster and more effective.11 However, while humoral immunity may provide long-lasting, antibody-mediated protection against future infections or severe disease caused by the same influenza strain, antibody directed against HA viral surface glycoprotein will often fail to neutralise influenza A viruses that have drifted, or influenza viruses of other types and subtypes.12 By contrast, antibodies against NA are not neutralising and limit infection and severity of illness by inhibiting NA and the release of new virions.13
The role of bacterial infections in severe influenza
In 1918, influenza was assumed to be a bacterial disease. Indeed, Haemophillus Influenzae was described as the cause of influenza before influenza viruses were first isolated and characterized. The interaction between influenza and bacterial co-pathogens is of more than academic interest: most patients presenting with severe influenza are given antibiotics. It would be a brave clinical decision to withhold antibiotics, faced with a febrile patient with declining respiratory function and pulmonary consolidation. Less virulent seasonal influenza viruses also predispose to secondary infection, but the 1918 virus resulted in a remarkable pandemic of lethal bacterial pneumonia.14
The question of the role of secondary bacterial pneumonia in the 1918-19 pandemic has been repeatedly examined. For example, Morens et al.15 reviewed 1,539 publications with pathology and/or microbiology results from autopsy investigations of 8398 individuals across 15 countries and examined histological slides re-cut from lung tissue blocks obtained 90 years earlier during autopsies of 58 individuals, concluding that bacterial pneumonia was indeed present in most or all of those who died. The types of bacteria causing pneumonia were diverse, reflecting the typical bacteria present in the upper respiratory tract, and Streptococcus pneumoniae was one of the more commonly identified bacterial species. Morens et al.15 concluded that susceptibility to secondary bacterial pneumonia resulted from defective antibacterial defences caused by viral infection, and that prevention and treatment of secondary bacterial pneumonia (including stockpiling of antibiotics and bacterial vaccines) should be included in pandemic plans. In addition to Streptococcus pneumoniae, Staphylococcus aureus and Streptococcus pyogenes are commonly isolated secondary bacterial infections in influenza and associated with increased risk of mortality.16,17
Determining the mechanism by which influenza predisposes to secondary infection18,19 and deciding who should be treated for bacterial superinfections has been the focus of considerable effort. Interestingly, influenza infection (with a live attenuated vaccine) has been reported to permit elevated colonization by Streptococcus pneumoniae.20 By comparison, antibiotic induced microbiome dysbiosis has been reported to impede the antibody response to influenza vaccination,21 indicating that commensal microbes normally enhance anti-influenza immunity, but can initiate bacterial pneumonia if the inflammatory response to influenza is exaggerated. To address this issue, the MOSAIC consortium investigated which aspects of the host response and pathogen might together explain the severity of disease. This study linked extensive clinical and investigational data to viral isolates, bacterial profiles, whole blood mRNA signatures, local and systemic immune mediator concentrations in 131 adults hospitalised with influenza. mRNA ‘signatures’ (groups of activated genes) reflecting IFN-related antiviral pathways predominated up to day 4 of illness in cases not requiring mechanical ventilatory support; in those needing mechanical ventilation an inflammatory, neutrophil and cell stress/death (‘bacterial’) pattern was seen, even early in disease. Identifiable bacterial co-infection was not necessary for this ‘bacterial’ signature but could enhance its development, while attenuating the early ‘viral’ signature.5 These findings show the importance of timing and severity in the interpretation of host responses to severe influenza.
Influenza infections in asthmatics
We recently completed a reanalysis of the MOSAIC cases to determine the differential pathogenesis of influenza H1N1pdm09 in adults with or without asthma, the most prevalent pre-existing medical condition (present in 30% of cases). In this subgroup, there was an excess of females (70% of asthmatics vs. 39% of non-asthmatics). The disease was less severe, with shorter length of stay (8.3 vs. 15.3 days) and lower mortality (3% vs. 11%). In the first 24 h, patients with asthma had reduced systemic markers of inflammation with lower serum IL-6, TNF, CXCL-8, 9, IL-10, IL-17 and CCL-2. There was no difference in terms of virus titres. Notably, those with asthma did not demonstrate a heightened type 2 immune signature and (although peripheral blood eosinophils and atopic status were not recorded) there was no difference in serum total IgE, periostin, blood eosinophil gene expression, or nasal IL-13, IL-4 or IL-5.22 Thus, those with asthma were more likely to present because of their airway symptoms as opposed to the seriousness of the systemic influenza infection. For those without asthma, a greater systemic immune response is associated with worse outcomes.
Pandemic influenza
It is generally agreed that the question is not ‘if’ but ‘when’ there will be another influenza pandemic. The pattern of threat varies around the globe, influenza A viruses circulating not only in human populations but also in pigs and in many species of domestic birds, shore birds and waterfowl.23 Indeed, the principal reservoirs of influenza A viruses are aquatic birds, rather than humans and other animals. The ability of influenza A to mutate and jump within and between species and the potential of avian, porcine and human strains to recombine give influenza A an extraordinary ability to exploit novel ecological niches.3,23
In addition to meeting the epidemiological criteria for a pandemic, novel pandemic influenza A viruses differ from season influenza A viruses in terms of the composition of their major surface antigens. Pandemic influenza A viruses arise as a result of antigenic shift; that is, the expression of novel HA and/or NA proteins.3 Such a dramatic change results in viruses that are distinct from existing, circulating seasonal influenza A viruses, against which pre-existing antibodies might offer little protection. As antigenic shift is difficult to predict, so too is the ability to prepare pandemic vaccines in advance of an emerging pandemic virus. By contrast, established seasonal influenza A viruses undergo a process of regular antigenic drift, whereby the major antigens do not change abruptly and there is no change in the influenza A subtype, but cumulative viral genetic changes result in antigenic differences that are sufficient to facilitate escape from existing immune responses (particularly antibody based immunity) acquired from previous infections or immunisation.24,25 This antigenic drift is the reason that the composition of seasonal influenza vaccines needs to be reviewed, and typically updated, on an annual basis.25
Epidemics of influenza-like illness were described by Hippocrates in ancient Greece, but the first convincing flu pandemic was described in 1510.26 The 2009 influenza (pH1N1) pandemic resulted in generally mild disease but still caused an estimated 250,000–500,000 additional deaths during the first 12 months of global circulation.27,28 Whereas seasonal influenza commonly causes severe disease in the old and infirm, pH1N1 disease mostly affected infants and younger adults, sometimes presenting as viral pneumonia occasionally complicated by multi-organ failure.29 The explanation for the age-associated pattern is most likely to be an absence of influenza subtype-specific immunity in H1N1-naïve populations. Other groups identified as being at increased risk of severe pH1N1 disease include pregnant women and obese individuals.30,31,32
Despite advances in science and medicine, the prodigious potential of influenza to mutate and spread remains a threat to human and animal health. The wide range of hosts provides influenza A viruses with greater chances of genetic re-assortment, leading to the emergence of zoonotic strains with the potential to spread to human populations. This has occurred on several occasions, but usually with limited ability to transmit person to person. Of greatest current concern is the continued excursions of avian H7N9 virus33,34 into those in contact with poultry. So far, this particularly dangerous virus has not shown sustained human transmission, but might mutate and acquire this ability in the future.
It has been suggested that severe pandemic influenza may in part result from an over-exuberant host reaction to infection (sometimes termed ‘cytokine storm’), but is also driven by a high viral load in affected persons.35 The exact chain of events that leads to severe disease has not been defined, but in one study negative influenza serology 4 days after the onset of influenza symptoms predicted death from fulminant influenza. The authors speculate that this might reflect trapping of anti-H1N1 antibodies in immune complexes in the lungs, associated with poor specific helper T-cell responses and high levels of IL-6 and IL-10 levels in both plasma and lung fluid.36
Seasonal influenza
The global impact of seasonal influenza is hard to estimate because so many cases are never identified, but it is estimated to contribute 30% of the total burden of infectious disease in Europe- more than any other single infection.37 A high proportion of deaths resulting from influenza are likely to be ascribed to other causes including COPD, bacterial pneumonia, decompensated heart disease or stroke.
To give an example of the hidden impact of influenza, national surveillance data from Scotland were used to identify adults suffering myocardial infarction or stroke from 2004 to 2014 and these events linked to records of laboratory-confirmed respiratory infections. This study showed greatly increased rates of myocardial infarction in the week after Streptococcus pneumoniae and influenza virus infection with adjusted incidence ratios (IRs) 5.98 and 9.80, respectively. Rates of stroke were also elevated, with IRs 12.3 and 7.82, respectively.38 A recently reported, self-controlled case series study examined hospitalisations for acute myocardial infarction that occurred within 12 months before and 12 months after a positive diagnostic test result for influenza virus infection. This study showed an increased rate of acute myocardial infarction in the seven days following diagnosis of influenza, which was six times greater than the rate during the control intervals.39 Proposed mechanisms for the observed increases in acute cardiovascular events include the systemic inflammatory response, increased physiological demands and pro-thrombotic states that can be expected in acute infections, including influenza.40
Sophisticated modelling techniques have been used to calculate influenza-associated excess mortality rates (EMR) covering for 57% of the global population. The estimated mean annual influenza-associated respiratory EMR ranges from 0·1 to 6·4 per 100 000 individuals for people younger than 65 years, 2·9 to 44·0 per 100,000 individuals for people aged between 65 and 74 years, and 17·9 to 223·5 per 100,000 for people older than 75 years. It is estimated that between 291,000 and 646,000 seasonal influenza-associated respiratory deaths occur annually around the globe, the highest mortality rates being in sub-Saharan Africa (2·8–16·5 per 100,000 individuals), southeast Asia (3·5–9·2 per 100,000 individuals), and among people aged 75 years or older (51·3–99·4 per 100,000 individuals).41 It seems certain that influenza is associated with a burden of disease and mortality beyond those cases where influenza is identified as a primary cause.
Influenza vaccines
Statistical modelling techniques are used to analyze global cycles of influenza activity in order to decide the best approach to vaccination in different countries. Current strain selection and vaccine distribution approaches are designed for countries with typical Northern and Southern Hemisphere single-peak influenza seasons. However, in countries closer to the equator influenza vaccination may need to be year-round, even at times when vaccines are not in production for the more temperate zones.42 This provides a material challenge in tropical and subtropical countries where influenza seasonality is less well defined. Current approaches to the development of influenza vaccines are shown in Fig. 3.
a Structure of influenza virus. HA: viral hemagglutinin, NA: viral neuraminidase; b Inactivated vaccines. Virus produced by culturing in chicken eggs or by cell culture in animal cells. HA/NA from selected influenza A strains reassorted with high growth egg-adapted virus. These vaccines are widely used in those >6 months of age, in pregnant women and those with chronic health problems that increase the risk of severe influenza. Higher dose of antigens and/or addition of adjuvant may be used to improve immunogenicity, particularly in those >65 y. Parenteral route of administration inducing relatively poor levels of mucosal immunity. Propagation of vaccine strains in eggs may introduce genetic changes that decrease vaccine effectiveness in man. c Live attenuated influenza vaccines (LAIV). These are cold adapted, therefore replicating poorly at 37 °C. Internal virus genes carry multiple attenuating temperature sensitive and cold adapting mutations virus with regularly updated HA/NA genes inserted by re-assortment or recombinant techniques. Approved for healthy people aged 2–49. Not suitable for those with immunodeficiency, in very young children or asthmatics due to possible risk of inducing wheeze. More closely mimics natural infection and may induce broader protection. Marketed as Flumist or Fluenz Tetra; d Recombinant vaccine production. HA gene from a wild-type flu virus in vector virus that grows in insect cells. HA protein is extracted and purified from cell culture. Manufacturing does not require propagation of influenza virus in chicken eggs, allowing potentially faster start-up for production. May be used for a novel pandemic vaccine manufacture, but may have shorter shelf-live. Used in the USA for those aged over 18 y (Flublok Quadrivalent).
Twice a year, the process of identifying the next vaccine strains to be recommended by the World Health Organization starts with identifying which viral HA and NA variants are emerging that may have novel antigenic qualities and thus evade immunity generated in the human population by last year’s strains and vaccines. This estimation of the strains and HA and NA variants that are considered likely to circulate is an imperfect art, resulting in poor strain matches between vaccines and circulating viruses and lower vaccine effectiveness in some years. The virulent 1918 virus was an H1N1 subtype and apparently highly pathogenic; by contrast, the 2009 H1N1 pandemic caused a great deal of mild and often unapparent disease.43
In the winter of 2017–18, H3N2 (a descendent of the 1968 pandemic) caused a great deal of severe disease in some areas, as it has done in some previous years. Unlike vaccination, natural influenza induces a high proportion of NA-reactive B cells. Such antibodies are thought to decrease disease severity by inhibiting NA and the release of new virions.13 Human antibodies binding NA epitopes bind to many influenza virus strains and show prophylactic and therapeutic efficacy in mice, but may not offer sterilizing immunity.44
The most commonly used current influenza vaccines fall into two broad groups: trivalent or quadrivalent inactivated vaccines (injected purified influenza surface protein preparations), and the live attenuated (quadrivalent) inhaled influenza vaccines (LAIV) that have been used in the USA in the past, but more recently introduced for childhood vaccination in the UK and other countries.28,45 Within the inactivated vaccines further differences exist, including the method used to propagate the vaccine strain (for example egg-based versus cell-based culture), the addition of immunological adjuvants, the amounts of antigens and adjuvants, and the routes of administration (intramuscular versus intradermal) (Fig. 3b). Although the most commonly used LAIVs are quadrivalent, trivalent LAIVs also exist45 (Fig. 3c). In the USA, authorities resumed the recommendation to use quadrivalent LAIV during the 2018-2019 influenza season, along with other vaccines, following earlier concerns about LAIV effectiveness in children and adolescents.46,47 In addition, recombinant vaccines have been developed, which combine HA genes from circulating influenza viruses with genes from viruses that grow well in insect cells, which avoids the need for egg-based propagation48 (Fig. 3d). Currently, only one recombinant seasonal vaccine is approved for use in the USA.
The main focus of vaccination is to induce specific immunity in the form of antibody against the influenza surface proteins antigens HA and NA. Currently available vaccines utilize HA, however. This immunity is conveniently measured as haemagglutination inhibition (HAI) for those viruses that exhibit haemagglutination. In addition, LAIV induces immune responses to NA as well as HA, local IgA and influenza specific resident memory CD4+ and CD8+ T cells11,49,50 that may contribute to local mucosal defences. These forms of immunity may be more important than the induction of systemic antibody, a current regulatory requirement for conventional parenteral influenza vaccines. Methods of vaccine production are discussed in more detail elsewhere.51
Variants of influenza surface proteins arise because of changes of the amino acid sequences driven by immunity to prior circulating strains and by adaptation to the host. Growing influenza virus in eggs in order to make the seasonal vaccine can change the HA protein that mediates receptor binding because of adaptation to avian cells. Small modifications in HA sequence can cause significant antigenic changes and decrease vaccine effectiveness, particularly with influenza A (H3N2) viruses, even when the vaccine strain closely matches that of the circulating virus. For example, a recent study by Zost et al.52 highlighted a particular egg-adaptive mutation (T160K) that seems to have contributed to low vaccine effectiveness during the 2016–2017 influenza season. T160K deletes a glycosylation site that is critical for both ferret and human antibody induction. Alternatives to egg-based propagation of vaccine strains, such as recombinant techniques, have the potential to overcome this problem.
The UK has mainly used injectable subunit vaccines in adults, but recently introduced LAIV for children. The injectable vaccines induce antibody to the chosen strains of flu but give little or no protection against other strains of influenza.45 The protection they induce relies on the accuracy of strain selection and varies from year to year. In addition, the injectable form can contain three virus strains or four; the latter (quadrivalent) targets two influenza A viruses and two influenza B varieties, which again change over time.25 LAIV has a greater potential to induce mucosal immunity and to protect against poorly matched strains of flu,47,53 but the problem with all existing influenza vaccines is that they have to be changed each year to match the expected circulating strains and do not provide protection against novel or zoonotic strains (which includes some very dangerous viruses which may be lethal for a high proportion of those infected).25,45
An additional level of complexity is the emerging understanding of ‘original antigenic sin’ in the antibody response to influenza vaccines and viruses. The principle underlying this idea is that antibody response to a first influenza infection biases recall responses to subsequent influenza infections (even of different antigenic types). The concept first gained traction in the 1950s54,55 but had gained recent momentum with reports demonstrating broadly convergent antibody responses to H1N1 and H3N2 infections.56,57 Similarly, infections in childhood with H3 subtype viruses offers some protection against severe H7 subtype infections (since both are HA group 2 viruses), indicating that cross-protection does occur but is dependent on an individual’s first exposure.58 This concept offers insights to our immunity to influenza and might also be harnessed for the design of next-generation vaccines.59
There are intensive efforts to develop vaccines that provide broad, long-lasting protection against known and (yet) unknown strains of influenza. The commercial argument for an improved vaccine has limited traction, most of the drive coming from public health, government laboratories (especially in the USA), academia and from charitably funded laboratories. The goal is to develop an effective “broadly-reactive” or “universal” influenza vaccine capable of conferring protection against both seasonal and newly emerging pre-pandemic strains.60
A number of approaches are being taken to develop universal influenza vaccines, including the induction of cross-reactive (CD8+) T cells and those that induce antibodies to the head domain of HA.61 Novel antigen delivery systems and vaccination strategies include the use of CpG-adjuvanted peptide vaccines to provide heterosubtypic protection by inhibiting regulatory T cell (Treg) development, thus enhancing T-cell immunity.62
It is also possible that greater understanding of the way that viruses manipulate host Type I interferon systems63 might lead to design of vaccines of enhanced immunogenicity. For example, quantitative high-throughput genomics has recently been applied to discovering regions of the influenza A genome that affect replication fitness and IFN sensitivity. By building viruses incorporating eight IFN-sensitive mutations, novel vaccine candidates were produced that are highly attenuated in IFN-competent hosts but able to induce transient IFN responses with robust humoral and cellular immunity that provides protection against homologous and heterologous viral challenges.64
Next-generation vaccines may primarily aim to induce sterilizing immunity. However, they may also aim to reduce the severity of disease, lessen complications, accelerate recovery and reduce shedding of seasonal (or pandemic) influenza viruses of diverse types, which may not require sterilizing immunity. Novel approaches to design of novel viral vector systems may be complemented by ‘tuned’ adjuvants, or designer chimeric proteins optimised for the induction of specific humoral or cell-mediated immune responses. In addition, monoclonal antibodies or cocktails of antibodies might be developed for passive prophylaxis and treatment.65
Antiviral drugs
With antiviral drugs, a key issue is the timing of the intervention. There is little doubt that early treatment greatly reduces the frequency and severity of disease, but patients with influenza rarely present within the first 2 days of disease onset. More usually, cases are seen on day 4 or beyond, at which time antivirals may have a relatively small effect.66,67
WHO guidelines recommend antiviral treatment in uncomplicated influenza in those with risk factors for complicated illness, as well as treatment of patients with severe or progressive clinical presentations of suspected or confirmed influenza.68 Neuraminidase inhibitors (NAIs) are most often used; although adamantanes (amantadine and rimantadine) and ribavirin have activity against some influenza viruses, their use is limited by side-effects and antiviral resistance. Influenza B viruses are generally resistant to adamantanes,69 as are currently circulating seasonal influenza A viruses.
Oseltamivir and zanamivir are the two major NAIs currently in clinical use in most countries. Both target the NA surface protein, inhibiting cleavage of viral HA from sialic acid residues expressed on the cell surface.70 Oseltamivir is active orally (or via a nasogastric tube) and zanamivir is normally administered via inhalation as a dry powder (or as an intravenous infusion in certain circumstances). Alternatives are available in some countries including intravenous peramivir and the long-acting inhaled NAI, laninamivir,71 and Baloxavir marboxil, the most recent antiviral to be approved. Baloxavir is an oral, single-dose, cap-dependent endonuclease that inhibits influenza polymerase, thus interrupting the synthesis of new virions. The addition of an antiviral that acts in a different way to NAIs is useful, but there is already evidence of decreased susceptibility to baloxavir emerging in clinical trials.72 Since early treatment may prevent progression to severe illness,73 guidelines generally recommend empirical antiviral treatment (oseltamivir or zanamivir) when influenza is suspected and while awaiting laboratory confirmation.74,75 Molecular tests for influenza viruses, typically PCR detection methods, are commonly used in hospital and tend to perform better than non-molecular rapid diagnostic tests. Point of care molecular platforms mean that diagnostics are now available for use in clinical areas, in addition to laboratory-based diagnostic testing. Regardless of the methods used, high quality sampling is required. This includes obtaining lower respiratory tract samples when there is evidence of lower respiratory tract disease, since lower respiratory tract samples may be positive when upper respiratory tract samples are negative.76
However, there is controversy surrounding the use of antiviral drugs. A Cochrane review suggested that oseltamivir reduces duration of symptoms by 21 h in adults with uncomplicated seasonal influenza, with no statistical evidence of an effect on subsequent hospitalisation rates.77 By contrast, a comprehensive meta-analysis of the effects of oseltamivir in seasonal influenza concluded that treatment led to a 28% reduction in lower respiratory tract complications requiring antibiotic treatment and an even greater effect in those with laboratory-confirmed influenza.78 A meta-analysis of antiviral treatment in 29,234 adult hospitalised cases (78 studies, 2009–2011, predominantly pandemic H1N1 virus) found that NAI treatment was associated with a reduction in mortality, especially in those treated within two days of symptom onset. However, even delayed NAI treatment appears to be better than no treatment (especially in critically ill patients). Evidence of benefit is weaker in children,79 but a more recent systematic review of individual data from studies of children showed that oseltamivir reduces the duration of illness by an average of 17.6 h, with a greater effect in children with asthma (duration reduced by 29.9 h).80
Given the evidence that an exuberant inflammatory response contributes to the pathogenesis of severe influenza with pneumonitis and/or acute respiratory distress syndrome,5,81 it seems appropriate to consider using immunomodulatory agents82 in addition to targeting viral replication. Such interventions have shown potential benefits in vitro, in observational studies or in animal models.83,84 Proposed approaches include the use of macrolides85,86 and statins87 but the clinical benefits of immunomodulation have been hard to demonstrate.88 Corticosteroid treatment of hospitalised patients with influenza is generally to be avoided.89,90
Conclusions
Influenza viruses hold a unique position in the landscape of human pathogens, exceptional in the ease with which they transmit and the severity of their impact on an unlucky few. Great progress has been made in understanding influenza viruses, the way that they mutate and spread, the effects they have on the host and the ways that the host immune system responds.
However, many significant challenges remain. Current vaccines are largely limited to offering homosubtypic immunity, depend on accurate forecasting of anticipated seasonal strains and offer little protection against emergent pandemic viruses. More effective vaccines that provide heterosubtypic (universal) and long-lasting protection are needed; great strides have been made in these directions, but such vaccines remain elusive. The advent of antimicrobial drugs has greatly alleviated influenza mortality, but rising rates of resistance against frontline antibiotics challenge our ability to combat severe disease, particularly bacterial co-infections.
Armed with greater knowledge, influenza might be finally defeated. But, for now, influenza remains an ever-present cause of disease and death around the globe.
References
Ahmed, R., Oldstone, M. B. & Palese, P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat. Immunol. 8, 1188–1193 (2007).
WHO. Virology of Influenza. [cited 28 Oct 2019] http://www.euro.who.int/en/health-topics/communicable-diseases/influenza/data-and-statistics/virology-of-human-influenza.
Paules, C. & Subbarao, K. Influenza. Lancet 390, 697–708 (2017).
Chow, K. T., Gale, M. Jr & Loo, Y. M. RIG-I and other RNA sensors in antiviral immunity. Annu Rev. Immunol. 36, 667–694 (2018).
Dunning, J. et al. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nat. Immunol. 19, 625–635 (2018).
Cole, S. L. & Ho, L. P. Contribution of innate immune cells to pathogenesis of severe influenza virus infection. Clin. Sci. (Lond.) 131, 269–283 (2017).
Newton, A. H., Cardani, A. & Braciale, T. J. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin Immunopathol. 38, 471–482 (2016).
Grant, E. J., Quinones-Parra, S. M., Clemens, E. B. & Kedzierska, K. Human influenza viruses and CD8(+) T cell responses. Curr. Opin. Virol. 16, 132–142 (2016).
La Gruta, N. L. & Turner, S. J. T cell mediated immunity to influenza: mechanisms of viral control. Trends Immunol. 35, 396–402 (2014).
Koutsakos, M. et al. Circulating TFH cells, serological memory, and tissue compartmentalization shape human influenza-specific B cell immunity. Sci. Transl. Med. 10, eaan8405 (2018).
Zens, K. D., Chen, J. K. & Farber, D. L. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 1, e85832 (2016).
Li, C. K., Rappuoli, R. & Xu, X. N. Correlates of protection against influenza infection in humans–on the path to a universal vaccine? Curr. Opin. Immunol. 25, 470–476 (2013).
Marcelin, G., Sandbulte, M. R. & Webby, R. J. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev. Med. Virol. 22, 267–279 (2012).
Oxford, J. S. & Gill, D. Unanswered questions about the 1918 influenza pandemic: origin, pathology, and the virus itself. Lancet Infect. Dis. 18, e348–e354 (2018).
Morens, D. M., Taubenberger, J. K. & Fauci, A. S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970 (2008).
Chertow, D. S. & Memoli, M. J. Bacterial coinfection in influenza: a grand rounds review. JAMA 309, 275–282 (2013).
Finelli, L. et al. Influenza-associated pediatric mortality in the United States: increase of Staphylococcus aureus coinfection. Pediatrics 122, 805–811 (2008).
Robinson, K. M., Kolls, J. K. & Alcorn, J. F. The immunology of influenza virus-associated bacterial pneumonia. Curr. Opin. Immunol. 34, 59–67 (2015).
Roquilly, A. et al. Local modulation of antigen-presenting cell development after resolution of pneumonia induces long-term susceptibility to secondary infections. Immunity 47, 135–147 (2017). e135.
Jochems, S. P. et al. Inflammation induced by influenza virus impairs human innate immune control of pneumococcus. Nat. Immunol. 19, 1299–1308 (2018).
Hagan, T. et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178, 1313–1328 (2019). e1313.
Jha, A. et al. Patterns of systemic and local inflammation in patients with asthma hospitalised with influenza. Eur. Respir. J. 54, 1900949 (2019).
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. & Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 56, 152–179 (1992).
Gething, M. J., Bye, J., Skehel, J. & Waterfield, M. Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature 287, 301–306 (1980).
Treanor, J. Influenza vaccine–outmaneuvering antigenic shift and drift. N. Engl. J. Med. 350, 218–220 (2004).
Morens, D. M., North, M. & Taubenberger, J. K. Eyewitness accounts of the 1510 influenza pandemic in Europe. Lancet 376, 1894–1895 (2010).
Dawood, F. S. et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect. Dis. 12, 687–695 (2012).
Openshaw, P. J. & Dunning, J. Influenza vaccination: lessons learned from the pandemic (H1N1) 2009 influenza outbreak. Mucosal Immunol. 3, 422–424 (2010).
Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med 362, 1708–1719 (2010).
Myles, P. R. et al. Predictors of clinical outcome in a national hospitalised cohort across both waves of the influenza A/H1N1 pandemic 2009-2010 in the UK. Thorax 67, 709–717 (2012).
Segaloff, H. E. et al. The impact of obesity and timely antiviral administration on severe influenza outcomes among hospitalized adults. J. Med. Virol. 90, 212–218 (2018).
Short, K. R., Kedzierska, K. & van de Sandt, C. E. Back to the future: lessons learned from the 1918 influenza pandemic. Front Cell Infect. Microbiol 8, 343 (2018).
Dortmans, J. C. et al. Adaptation of novel H7N9 influenza A virus to human receptors. Sci. Rep. 3, 3058 (2013).
Uyeki, T. M. & Cox, N. J. Global concerns regarding novel influenza A (H7N9) virus infections. N. Engl. J. Med. 368, 1862–1864 (2013).
Peiris, J. S., Cheung, C. Y., Leung, C. Y. & Nicholls, J. M. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol. 30, 574–584 (2009).
Guihot, A. et al. Low titers of serum antibodies inhibiting hemagglutination predict fatal fulminant influenza A(H1N1) 2009 infection. Am. J. Respir. Crit. Care Med 189, 1240–1249 (2014).
Cassini, A. et al. Impact of infectious diseases on population health using incidence-based disability-adjusted life years (DALYs): results from the Burden of Communicable Diseases in Europe study, European Union and European Economic Area countries, 2009 to 2013. Euro. Surveill. 23, 17-00454 (2018).
Warren-Gash, C., Blackburn, R., Whitaker, H., McMenamin, J. & Hayward, A. C. Laboratory-confirmed respiratory infections as triggers for acute myocardial infarction and stroke: a self-controlled case series analysis of national linked datasets from Scotland. Eur. Respir. J. 51, 1701794 (2018).
Kwong, J. C., Schwartz, K. L. & Campitelli, M. A. Acute myocardial infarction after laboratory-confirmed influenza infection. N. Engl. J. Med. 378, 2540–2541 (2018).
Warren-Gash, C., Smeeth, L. & Hayward, A. C. Influenza as a trigger for acute myocardial infarction or death from cardiovascular disease: a systematic review. Lancet Infect. Dis. 9, 601–610 (2009).
Iuliano, A. D. et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300 (2018).
Newman, L. P., Bhat, N., Fleming, J. A. & Neuzil, K. M. Global influenza seasonality to inform country-level vaccine programs: An analysis of WHO FluNet influenza surveillance data between 2011 and 2016. PLoS ONE 13, e0193263 (2018).
Hayward, A. C. et al. Comparative community burden and severity of seasonal and pandemic influenza: results of the Flu Watch cohort study. Lancet Respir. Med. 2, 445–454 (2014).
Chen, Y. Q. et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173, 417–429 (2018). e410.
Treanor, J. J. CLINICAL PRACTICE. Influenza vaccination. N. Engl. J. Med. 375, 1261–1268 (2016).
Committee On Infectious, D. Recommendations for prevention and control of influenza in children, 2018–2019. Pediatrics 142 (2018).
Singanayagam, A., Zambon, M., Lalvani, A. & Barclay, W. Urgent challenges in implementing live attenuated influenza vaccine. Lancet Infect. Dis. 18, e25–e32 (2018).
Treanor, J. J. et al. Protective efficacy of a trivalent recombinant hemagglutinin protein vaccine (FluBlok(R)) against influenza in healthy adults: a randomized, placebo-controlled trial. Vaccine 29, 7733–7739 (2011).
Aljurayyan, A. et al. Activation and induction of antigen-specific T follicular helper cells play a critical role in live-attenuated influenza vaccine-induced human mucosal anti-influenza antibody response. J. Virol. 92, e00114-18 (2018).
Ambrose, C. S., Wu, X., Jones, T. & Mallory, R. M. The role of nasal IgA in children vaccinated with live attenuated influenza vaccine. Vaccine 30, 6794–6801 (2012).
Fraser, C. S., Jha, A. & Openshaw, P. J. Vaccines in the prevention of viral pneumonia. Clin. Chest Med. 38, 155–169 (2017).
Zost, S. J. et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl Acad. Sci. USA 114, 12578–12583 (2017).
Iwasaki, A. Exploiting Mucosal Immunity for Antiviral Vaccines. Annu Rev. Immunol. 34, 575–608 (2016).
Francis, T. Jr. Influenza: the new acquayantance. Ann. Intern. Med. 39, 203–221 (1953).
Kim, J. H., Skountzou, I., Compans, R. & Jacob, J. Original antigenic sin responses to influenza viruses. J. Immunol. 183, 3294–3301 (2009).
Nachbagauer, R. et al. Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nat. Immunol. 18, 464–473 (2017).
Rajendran, M. et al. Analysis of anti-influenza virus neuraminidase antibodies in children, adults, and the Elderly by ELISA and enzyme inhibition: evidence for original antigenic Sin. mBio 8, e02281-16 (2017).
Gostic, K. M., Ambrose, M., Worobey, M. & Lloyd-Smith, J. O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726 (2016).
Henry, C., Palm, A. E., Krammer, F. & Wilson, P. C. From original antigenic sin to the universal influenza virus vaccine. Trends Immunol. 39, 70–79 (2018).
Paules, C. I., Sullivan, S. G., Subbarao, K. & Fauci, A. S. Chasing seasonal influenza—the need for a universal influenza vaccine. N. Engl. J. Med. 378, 7–9 (2018).
de Vries, R. D., Altenburg, A. F. & Rimmelzwaan, G. F. Universal influenza vaccines, science fiction or soon reality? Expert Rev. Vaccines 14, 1299–1301 (2015).
Lin, P. H. et al. Vaccine-induced antigen-specific regulatory T cells attenuate the antiviral immunity against acute influenza virus infection. Mucosal Immunol. 11, 1239–1253 (2018).
Garcia-Sastre, A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 22, 176–184 (2017).
Du, Y. et al. Genome-wide identification of interferon-sensitive mutations enables influenza vaccine design. Science 359, 290–296 (2018).
Jackson, K. J. L. & Boyd, S. D. Broadening horizons: new antibodies against influenza. Cell 166, 532–533 (2016).
Beard, K. R., Brendish, N. J. & Clark, T. W. Treatment of influenza with neuraminidase inhibitors. Curr. Opin. Infect. Dis. 31, 514–519 (2018).
Katzen, J., Kohn, R., Houk, J. L. & Ison, M. G. Early oseltamivir after hospital admission is associated with shortened hospitalization: a 5-year analysis of oseltamivir timing and clinical outcomes. Clin. Infect. Dis. 69, 52–58 (2019).
WHO. Influenza (Seasonal) Ask the Expert: Influenza Q&A. 2018 06 Nov 2018 [cited 28 Oct 2019] https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal).
Hayden, F. G. & de Jong, M. D. Emerging influenza antiviral resistance threats. J. Infect. Dis. 203, 6–10 (2011).
Davies, B. E. Pharmacokinetics of oseltamivir: an oral antiviral for the treatment and prophylaxis of influenza in diverse populations. J. Antimicrob. Chemother. 65(Suppl 2), ii5–ii10 (2010).
Hayden, F. G. Newer influenza antivirals, biotherapeutics and combinations. Influenza Other Respir. Viruses 7(Suppl 1), 63–75 (2013).
Hayden, F. G. et al. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N. Engl. J. Med. 379, 913–923 (2018).
Aoki, F. Y. & Hayden, F. G. The beneficial effects of neuraminidase inhibitor drug therapy on severe patient outcomes during the 2009–2010 influenza A virus subtype H1N1 pandemic. J. Infect. Dis. 207, 547–549 (2013).
PHE. Influenza: treatment and prophylaxis using anti-viral agents. 2019 [cited 29 Oct 2019] https://www.gov.uk/government/publications/influenza-treatment-and-prophylaxis-using-anti-viral-agents.
CDC. Influenza Antiviral Medications. 2019 [cited 29 Oct 2019] https://www.cdc.gov/flu/professionals/antivirals/index.htm.
Reddy, K. P., Bajwa, E. K., Parker, R. A., Onderdonk, A. B. & Walensky, R. P. Relationship between upper respiratory tract influenza test result and clinical outcomes among critically Ill influenza patients. Open Forum Infect. Dis. 3, ofw023 (2016).
Jefferson, T. et al. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. Cochrane Database System. Rev. CD008965 (2014).
Hernán, M. A. & Lipsitch, M. Oseltamivir and risk of lower respiratory tract complications in patients with flu symptoms: a meta-analysis of eleven randomized clinical trials. Clin. Infect. Dis. 53, 277–279 (2011).
Muthuri, S. G. et al. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir. Med. 2, 395–404 (2014).
Malosh, R. E. et al. Efficacy and safety of oseltamivir in children: systematic review and individual patient data meta-analysis of randomized controlled trials. Clin. Infect. Dis. 66, 1492–1500 (2018).
de Jong, M. D. et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12, 1203–1207 (2006).
Teijaro, J. R. et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 146, 980–991 (2011).
Shale, M., Czub, M., Kaplan, G. G., Panaccione, R. & Ghosh, S. Anti-tumor necrosis factor therapy and influenza: keeping it in perspective. Ther. Adv. Gastroenterol. 3, 173–177 (2010).
Hussell, T., Pennycook, A. & Openshaw, P. J. Inhibition of tumor necrosis factor reduces the severity of virus-specific lung immunopathology. Eur. J. Immunol. 31, 2566–2573 (2001).
Bermejo-Martin, J. F., Kelvin, D. J., Eiros, J. M., Castrodeza, J. & Ortiz de Lejarazu, R. Macrolides for the treatment of severe respiratory illness caused by novel H1N1 swine influenza viral strains. J. Infect. Developing Ctries. 3, 159–161 (2009).
Hung, I. F. N. et al. Efficacy of clarithromycin-naproxen-oseltamivir combination in the treatment of patients hospitalized for influenza a(h3n2) infection: an open-label randomized, controlled, phase IIb/III trial. Chest 151, 1069–1080 (2017).
Havers, F. et al. Statin use and hospital length of stay among adults hospitalized with community-acquired pneumonia. Clin. Infect. Dis. 62, 1471–1478 (2016).
Laidler, M. R. et al. Statin treatment and mortality: propensity score-matched analyses of 2007-2008 and 2009-2010 laboratory-confirmed influenza hospitalizations. Open Forum Infect. Dis. 2, ofv028 (2015).
Zhang, Y. et al. Do corticosteroids reduce the mortality of influenza A (H1N1) infection? A meta-analysis. Crit. Care 19, 46 (2015).
Kim, S. H. et al. Corticosteroid treatment in critically ill patients with pandemic influenza A/H1N1 2009 infection: analytic strategy using propensity scores. Am. J. Respir. Crit. Care Med. 183, 1207–1214 (2011).
Author information
Authors and Affiliations
Contributions
J.D., R.S.T., and P.O. drafted the manuscript, which was revised for intellectual content and approved by all authors. P.O. is the guarantor.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dunning, J., Thwaites, R.S. & Openshaw, P.J.M. Seasonal and pandemic influenza: 100 years of progress, still much to learn. Mucosal Immunol 13, 566–573 (2020). https://doi.org/10.1038/s41385-020-0287-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-020-0287-5
This article is cited by
-
Early mucosal events promote distinct mucosal and systemic antibody responses to live attenuated influenza vaccine
Nature Communications (2023)
-
Validation of Multi-epitope Peptides Encapsulated in PLGA Nanoparticles Against Influenza A Virus
Pharmaceutical Research (2023)
-
Narrative review on century of respiratory pandemics from Spanish flu to COVID-19 and impact of nanotechnology on COVID-19 diagnosis and immune system boosting
Virology Journal (2022)
-
Nanoparticle composite TPNT1 is effective against SARS-CoV-2 and influenza viruses
Scientific Reports (2021)
-
Clinical characteristics of Kawasaki disease and concurrent pathogens during isolation in COVID-19 pandemic
World Journal of Pediatrics (2021)