Abstract
The immune-regulatory microRNA miR-155 is reduced in recurrent miscarriage, suggesting that miR-155 contributes to immune tolerance in pregnancy. Here we show miR-155 is induced in the uterine mucosa and draining lymph nodes (dLN) during the female immune response to male seminal fluid alloantigens. Mice with null mutation in miR-155 (miR-155−/−) exhibited a reduced CD4+ T cell response after mating, with a disproportionate loss of CD25+FOXP3+ Treg cells. miR-155 deficiency impaired expansion of both peripheral and thymic Treg cells, distinguished by neuropilin-1 (NRP1), and fewer Treg cells expressed Ki67 proliferation marker and suppressive function marker CTLA4. Altered Treg phenotype distribution in miR-155−/− mice was confirmed by t-distributed neighbor embedding (tSNE) analysis. Fewer dendritic cells (DCs) and macrophages trafficked to the dLN of miR-155−/− mice, associated with lower CCR7 on DCs, and reduced uterine Ccl19 expression, implicating compromised antigen presentation in the stunted Treg cell response. miR-155−/− mice exhibited elevated susceptibility to inflammation-induced fetal loss and fetal growth restriction compared with miR-155+/+ controls, but outcomes were restored by transfer of wild-type Tregs. Thus miR-155 is a key regulator of immune adaptation to pregnancy and is necessary for sufficient Tregs to achieve robust pregnancy tolerance and protect against fetal loss.
Similar content being viewed by others
Introduction
An adaptive immune response to conceptus alloantigens occurs during early pregnancy in the uterine mucosa to establish and sustain immune tolerance for embryo implantation and pregnancy progression.1,2 Disruption of this immune response can alter the trajectory of placental and fetal development and reduce fertility and fecundity.1,2 In women, a dysregulated uterine immune response is implicated in common reproductive conditions, including recurrent implantation failure, recurrent miscarriage, and preeclampsia.3,4 In order to develop interventions to tackle these conditions, it is necessary to define the mechanisms underpinning adaptive immune tolerance in the uterus and the molecular regulators controlling quality and strength of the response.2
In particular, CD4+ regulatory T cells (Tregs) are essential for uterine tolerance of pregnancy.1,5 Tregs are defined by the expression of CD25 and transcription factor forkhead box P3 (FOXP3) and exhibit potent anti-inflammatory and immune-suppressive activity. They originate in the thymus (thymic Treg (tTreg)) or are converted from conventional T cells in the periphery (peripheral Treg (pTreg)). In mice, the two lineages can be distinguished by the expression of neuropilin-1 (NRP1). tTregs express high NRP16,7,8 and emerge from the thymus after TCR-driven selection, with stable suppressive function9 that is retained after antigen-driven activation and proliferation in certain peripheral tissues.10,11,12 pTregs are generated extrathymically from naive CD4 T cells responding to antigen stimulation in the presence of transforming growth factor beta (TGFβ) and interleukin (IL)-213 and are NRP1 negative or low (NRP−/lo is hereafter referred to as NRP1−).6,7,8
The uterine Treg cell population supports embryo implantation and pregnancy progression through suppressing adverse immune effector responses to fetal antigens, controlling local inflammatory activation, and promoting vascular adaptations required for robust placental development.14,15,16 Pregnancy tolerance involves both tTregs and pTregs, both of which undergo antigen-driven expansion in the uterus-draining para-aortic lymph nodes (dLN) during the days after conception,17,18 to generate an expanded Treg pool available for recruitment from peripheral blood into the decidua before embryo implantation.14,15 While tTreg make up the majority of the dLN and the uterine Treg population,19 extrathymically induced pTreg cells with reactivity to paternal alloantigens are also essential for optimal pregnancy success.20,21 Similarly in women, the pool of Treg cells expands during early pregnancy, with peripheral blood and uterine Tregs increasing over the course of the first trimester.22,23 Insufficient Tregs or impaired Treg function, with a counteractive increase in effector T cells (Teff cells), is a common feature in reproductive disorders and complications of human pregnancy,2,4 with unexplained infertility, recurrent miscarriage, preeclampsia, and preterm birth all associated with disrupted Treg cells.24,25,26,27
Experiments in mice show that antigen priming and expansion of the dLN Treg pool is initiated in the uterine mucosa and requires ovarian hormones,28 as well as paternal alloantigens and immune-regulatory signals TGFβ29 and ligands of Toll-like receptor 4,30 delivered in seminal fluid at coitus.17,18,31 These factors elicit a transient inflammation-like response in the endometrial lining of the uterus,32 characterized by extensive induction of pro-inflammatory cytokines and chemokines in the uterine endometrium.30 Elevated cytokines cause dendritic cells (DCs) and macrophages to be recruited into the endometrial stroma subjacent to the luminal epithelium,33 where they take up seminal fluid antigens, then emigrate to dLN, and induce alloantigen-reactive Treg cell populations to expand.34 Seminal fluid induces a similar immune response in women,3,35 and Treg cells located in the decidualized endometrium of early pregnancy exhibit fetal antigen specificity,23,24 implying a similar sequence of steps for Treg cell generation and recruitment in humans.
MicroRNA-155 (miR-155) is potently upregulated in T and B lymphocytes after antigen stimulation,36,37 as well as DCs and macrophages after Toll-like receptor ligation,38,39 with distinct lineage-specific effects on gene transcription and cell function.39,40 miR-155 influences phenotype commitment in CD4+ T cells, to play a pivotal role in shaping the balance between tolerance and immunity.36,41,42 Notably miR-155 is induced by FOXP3 transactivation to be expressed highly in Treg cells, and is required for normal thymic Treg cell generation.43,44 miR-155 maintains competitive fitness of Treg cells by targeting SOCS1 to allow IL-2-mediated induction of signal transducer and activator of transcription 5 (STAT5) transcription factor,41,44 but is not required for normal suppressive function.44,45 Conventional CD4+ T cells also express miR-155, with miR-155 expression levels influencing sensitivity to tTreg-mediated suppression.45 In innate immune cells, miR-155 facilitates acquisition of a pro-inflammatory phenotype in macrophages46,47 and influences DC trafficking, phenotype and function to regulate antigen presenting activity.39,48,49
miR-155 has comparable functions in mouse and human Treg cells50 and macrophages,51 and miR-155 is dysregulated in several inflammatory and autoimmune diseases.52,53,54 Altered miR-155 is reported in infertility associated with recurrent implantation failure,55 recurrent miscarriage,56 and preeclampsia,57 suggesting that miR-155 may modulate immune adaptation for pregnancy in women. Thus we hypothesized that miR-155 contributes to competent pregnancy tolerance through an impact on shaping the Treg response at conception. Utilizing mice with genetic miR-155 deficiency, we demonstrate that miR-155 is required for early expansion of the Treg cell pool, with consequences for Treg dynamics in later pregnancy. Impaired Tregs lead to increased susceptibility to inflammation-induced fetal loss in late gestation that is rescued by administration of Treg cells from wild-type miR-155-sufficient donors. Together, these data demonstrate a key role for miR-155 in maternal Treg generation at conception and setting the course of pregnancy tolerance.
Results
miR-155 is induced in the uterus and dLN in early pregnancy
To investigate the contribution of miR-155 to pregnancy tolerance, we first quantified miR-155 expression in the reproductive tract during the conception and embryo implantation phase, when the T cell response to paternal alloantigens is initiated. In the endometrial tissue lining the uterine mucosa, miR-155 was increased 6.2-fold at 36 h after mating with allogeneic males and remained elevated for at least 48 h (Fig. 1a), consistent with induction during the immune response to seminal fluid.2 Fluctuations in ovarian hormones over the 4-day mouse estrus cycle partially explain the elevated uterine miR-155, with a 1.7-fold increase over the pro-estrus to met-estrus transition in unmated cycling mice (Fig. 1b). Maximum miR-155 induction required contact with seminal fluid, and both the sperm and seminal plasma fractions contributed, since vasectomized males or seminal vesicle gland-excised males elicited a smaller increase (Fig. 1c). Male alloantigens were also required, since seminal fluid of allogeneic males elicited a stronger miR-155 response than that of seminal fluid of syngeneic males (Fig. 1d).
miR-155 expression was quantified by qPCR in uterus and dLN recovered from B6 mice at specific stages of the estrous cycle or after mating (8, 16, 36, 60, and 84 h pc). Data are miR-155 expression relative to U6snRNA reference in the uterine endometrium at estrus (est) and at 8–84 h following mating with intact BALB/c males (a); endometrium at pro-estrus (pro), estrus (est), met-estrus (met), and di-estrus (di) (b); endometrium at estrus (est) and on d3.5 pc following mating with BALB/c males surgically rendered seminal vesicle deficient and vasectomized (svx/vas), seminal vesicle deficient (svx), vasectomized (vas), or intact males (int) (c), and endometrium at estrus (est) and on d3.5 pc following mating with intact syngeneic (syn, B6) or allogeneic (allo, BALB/c) males (d), and dLN at estrus or 8–84 h following mating with intact BALB/c males (e). Symbols depict values from individual mice (n = 3–15/group) and mean ± SEM values are shown. Effects of mating and cycle stage were assessed by non-parametric Kruskal–Wallis test with post hoc Dunn’s multiple comparison test (a, b) or one-way ANOVA with Sidak t test (c–e) (*p < 0.05 compared to estrus; ǂp < 0.05 compared to the svx/vas, svx, and vas groups; ∧p < 0.05 compared to pro-estrus; #p < 0.05 compared to syngeneic).
Increased miR-155 expression in the uterus after mating was accompanied by similar changes in the dLN, the predominant site of T cell activation and proliferation after mating,34,58 where miR-155 expression was increased by 3.3-fold over estrus levels at 36 h after mating with allogeneic males and remained elevated for at least 48 h (Fig. 1e).
miR-155 deficiency reduces the dLN CD4+ Treg cell pool in early pregnancy
The role of miR-155 in the adaptive immune response to conception and pregnancy was investigated using mice with miR-155 null mutation.36 miR-155 was undetectable in the endometrium of unmated or mated miR-155−/− mice (Supplementary Fig. S1). Total CD4+ T cells and CD4+CD25+FOXP3+ Treg cells were assessed in dLN of pregnant miR-155−/− and miR-155+/+ mice at early, mid-, and late gestation on day (d) 3.5, d9.5 and d15.5 post-coitum (pc) and in unmated estrous mice (Fig. 2a). CD4+ T cell numbers were comparable in unmated miR-155−/− and miR-155+/+ mice, but the substantial rise in CD4+ T cells seen in miR-155+/+ mice on d3.5 pc (3.8-fold, consistent with previous studies19,58) failed to occur in miR-155−/− mice (Fig. 2b). Treg cells were particularly impacted, with a ~50% reduction in the proportion of Tregs among CD4+ T cells in miR-155−/− compared with miR-155+/+ mice before mating and over the course of gestation. While Tregs progressively increased as a proportion of CD4+ T cells after mating in miR-155+/+ mice, there was a smaller expansion in miR-155−/− mice (Fig. 2c). The increase in Treg cells was sustained into mid-gestation in miR-155+/+ mice, but no comparable expansion occurred in miR-155−/− mice, and their absolute numbers were reduced by 75%, 68%, and 77% at d3.5, d9.5 pc, and d15.5 pc, respectively (Fig. 2d).
CD4+ Treg cells in dLN recovered from miR-155+/+ (+/+) and miR-155−/− (−/−) female mice at estrus (est) or after mating (d3.5, 9.5, or 15.5 pc) were analyzed by flow cytometry for CD4, CD25, and FOXP3 expression. Representative flow cytometric plots show the relevant populations and proportion of cells (%) (a). Data are the number of CD4+ T cells (b), the percentage of CD4+CD25+FOXP3+ Treg cells within CD4+ T cells (c), and the number of CD4+CD25+FOXP3+ Treg cells (d), with symbols depicting values from individual mice (n = 7–15/group) and mean ± SEM shown. The effect of genotype at each time point and differences within genotype across the time course was assessed by a mixed-effects model with the Geisser–Greenhouse correction followed by Sidak multiple comparisons test (*p < 0.05 vs. same time point in miR-155+/+, τp < 0.05 difference attributable to mating, #p < 0.05 vs. estrus within genotype, ^p < 0.05 difference attributable to genotype).
Conversely, the proportion of CD4+Foxp3− Tconv cells among dLN CD4+ T cells was elevated, resulting in a 40–50% reduced Treg:Tconv ratio in miR-155−/− mice compared to miR-155+/+ mice over the course of pregnancy (Supplementary Fig. S2a–c). Unlike Tregs, the proportion of CD4+CD25+Foxp3− Teff cells among dLN CD4+ T cells was unchanged by miR-155 deficiency, resulting in a 30–50% reduced Treg:Teff ratio in miR-155−/− mice (Supplementary Fig. S2d–f).
In the spleens and mesenteric lymph nodes (MLNs), mating did not elicit changes in CD4+ T cells or Treg cells in either genotype. As in dLNs, the proportion of Tregs among CD4+ T cells was reduced at estrus and d3.5 pc in miR-155−/− mice compared with miR-155+/+ mice in these tissues (Supplementary Fig. S3).
Both thymic and peripheral Tregs are impacted by miR-155 deficiency
Additional markers were included to evaluate the effects of miR-155 deficiency on dLN Treg cell phenotype, including NRP1 to distinguish tTreg and pTreg cells,6,7,8 CTLA4, a marker of Treg-suppressive competence,59,60 and Ki67 to detect proliferating T cells. Both NRP1− pTreg and NRP1+ tTreg were reduced in miR-155−/− mice (Fig. 3a–d). In miR-155+/+ mice, both tTregs and pTregs contributed to expanded Treg cell number after mating, by increasing 4.5-fold and 7.4-fold, respectively. In miR-155−/− mice, there was no change in tTreg, and pTreg increased only 2.7-fold (Fig. 3e–g).
Total Treg cells, NRP1+ tTreg, and NRP1− pTreg cells in the dLN recovered from miR-155+/+ (+/+) and miR-155−/− (−/−) female mice at estrus (est) or after mating (d3.5 pc) were analyzed by flow cytometry for CD4, CD25, FOXP3, NRP1, CTLA4, and Ki67 expression. Representative flow cytometric plots show the relevant populations and proportion of cells (%) (a). Data are the percentage of CD4+CD25+FOXP3+ Treg cells within CD4+ T cells (b), the percentage of CD4+CD25+FOXP3+NRP1+ tTreg cells within CD4+ T cells (c), the percentage of CD4+CD25+FOXP3+NRP1− pTreg cells within CD4+ T cells (d), the number of CD4+CD25+FOXP3+ Treg cells (e), the number of CD4+CD25+FOXP3+NRP1+ tTreg cells (f), and the number of CD4+CD25+FOXP3+NRP1− pTreg cells (f), with symbols depicting values from individual mice (n = 8–12/group) and mean ± SEM shown. Differences between groups were assessed by one-way ANOVA with Sidak t test (*p < 0.05 vs. same time point in miR-155+/+, #p < 0.05 vs. estrus within genotype). The overall effect of genotype and mating was assessed by two-way ANOVA (∧p < 0.05 difference attributable to genotype; τp < 0.05 difference attributable to mating).
In addition, miR-155−/− mice had fewer CTLA4+ Tregs undergoing proliferation (Fig. 4a), due to the smaller Treg pool as opposed to any consistent change in the proportion expressing Ki67 or CTLA4 (Supplementary Fig. S4) or both markers (Fig. 4b–d). Double positive CTLA4+Ki67+ Tregs comprised ~20% of dLN Tregs in miR-155+/+ mice on d3.5 pc (Fig. 4b), reflecting a 12.2-fold increase in total CTLA4+Ki67+ Tregs compared to estrous (Fig. 4e). In contrast, total CTLA4+Ki67+ Tregs were reduced by 84% in miR-155−/− mice, after increasing only 3.9-fold after mating (Fig. 4b, e). CTLA4+Ki67+ cells increased similarly in tTreg (Fig. 4c, f) and pTreg cells (Fig. 4d, g) after mating in miR-155+/+ mice, but were fewer amongst both Treg subsets in miR-155−/− mice. Among CTLA4+ Tregs, CTLA4 mean fluorescent intensity (MFI) was slightly reduced in miR-155−/− mice, but FOXP3 MFI in Tregs was not substantially affected by genotype or mating (Supplementary Fig. S4).
CD4+ T cell CTLA4 and Ki67 expression in uterus-draining dLN recovered from miR-155+/+ (+/+) and miR-155−/− (−/−) female mice at estrus (est) or after mating (d3.5 pc) were analyzed by flow cytometry for CD4, CD25, FOXP3, NRP1, CTLA4, and Ki67 expression. Representative flow cytometric plots show the relevant populations and proportion of cells (%) (a). Data are the percentage of CD4+CD25+FOXP3+CTLA4+Ki67+ Treg cells within CD4+CD25+FOXP3+ Treg cells (b), the percentage of CD4+CD25+FOXP3+NRP1+CTLA4+Ki67+ tTreg cells within CD4+CD25+FOXP3+NRP1+ tTreg cells (c), the percentage of CD4+CD25+FOXP3+NRP1-CTLA4+Ki67+ pTreg cells within CD4+CD25+FOXP3+NRP1− pTreg cells (d), the number of CD4+CD25+FOXP3+CTLA4+Ki67+ Treg cells (e), the number of CD4+CD25+FOXP3+NRP1+CTLA4+Ki67+ tTreg cells (f), and the number of CD4+CD25+FOXP3+NRP1-CTLA4+Ki67+ pTreg cells (g), with symbols depicting values from individual mice (n = 9–12/group) and mean ± SEM shown. Differences between groups were assessed by non-parametric Kruskal–Wallis test with post hoc Dunn’s multiple comparison test (b, c) or one-way ANOVA with Sidak t test (d–g) (*p < 0.05 vs. same time point in miR-155+/+, #p < 0.05 vs. estrus within genotype). The overall effect of genotype and mating was assessed by two-way ANOVA (^p < 0.05 difference attributable to genotype; τp < 0.05 difference attributable to mating).
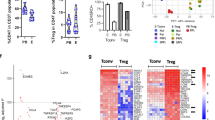
t-distributed stochastic neighbor embedding (tSNE) analysis confirms differential effects of miR-155 on distinct dLN tTreg subsets
tSNE was utilized as a complementary approach to investigate the impact of miR-155 deficiency on the dLN Treg cell response. tSNE utilizes an unsupervised algorithm to combine and integrate flow cytometric data, using all marker information to identify cell subpopulations in an unbiased way. Flow cytometric data on dLN CD4+ T cells from estrous and mated miR-155+/+ and miR-155−/− mice stained for CD4, FOXP3, CD25, NRP1, CTLA4, and Ki67 were concatenated and transformed into two-dimensional projections using the tSNE algorithm. Inspection of density and contour plots revealed distinct clusters, among which populations of FOXP3+ cells with the characteristics of tTreg and pTregs could be located (Fig. 5a–c). Cluster 1 corresponded to FOXP3+NRP1+ tTregs and cluster 2, an assembly of 4 smaller clusters, corresponded to FOXP3+NRP1− pTregs. The proportions of CD4+ T cells comprised by Tregs (cluster1+cluster 2) and specifically cluster 1 were fewer in miR-155−/− mice (Fig. 5d–f). In addition, a lower proportion of cluster 1 but not cluster 2 Treg cells expressed CTLA4 after mating (Fig. 5g, h). Z-score analysis of cluster 1 Treg phenotypes after mating in miR-155−/− mice suggested lower CTLA4, Ki67, FOXP3, and NRP1 than in miR-155+/+ mice (Fig. 5i), while CTLA4 and Ki67 were not obviously impacted in cluster 2 Treg cells (Fig. 5j).
CD4+ T cells from dLN of estrus and d3.5 pc mated mice were labeled to detect CD25, Ki67, NRP1, CTLA4, and FOXP3, and compensated.fcs files were concatenated by treatment group and transformed by the tSNE algorithm for FOXP3 (a) and NRP1 (b). FOXP3+NRP1+ (cluster 1) and FOXP3+NRP1− (cluster 2) were manually defined with the aid of contour plots (a, b) to approximate tTreg and pTregs, respectively (c). The percentage of FOXP3+ (cluster 1+2) within CD4+ T cells (d), the percentage of FOXP3+NRP1+ (cluster 1) within CD4+ T cells (e), and the percentage of FOXP3+NRP1− (cluster 2) within CD4+ T cells (f) are shown, with symbols depicting values from individual mice (n = 6–10/group) and mean ± SEM values. The proportions of FOXP3+NRP1+ (cluster 1) (g) and FOXP3+NRP1− (cluster 2) (h) comprising CTLA4+Ki67+, CTLA4+Ki67−, CTLA4-Ki67+, and CTLA4-Ki67− subsets, are shown. Effects on proportions due to mating and genotype were analyzed by Chi-square analysis. The average MFI expression of each phenotype marker in cluster 1 (i) and cluster 2 (j) at estrus and on d3.5 pc in miR-155+/+ and miR-155−/− mice is shown on a heat map of Z-score, where the average MFI is color coded and ranges from yellow (+1) to blue ( −1) according to the color scale. Differences between groups were assessed by non-parametric Kruskal–Wallis test with post hoc Dunn’s multiple comparison test (*p < 0.05 vs. same time point in miR-155+/+, #p < 0.05 vs. estrus within genotype). The overall effect of genotype and mating was assessed by two-way ANOVA (^p < 0.05 difference attributable to genotype; τp < 0.05 difference attributable to mating).
miR-155 deficiency alters DC populations in the uterus and dLN
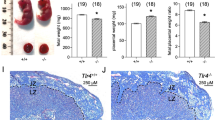
DCs drive expansion of dLN Treg cell populations in early pregnancy after they infiltrate from peripheral blood after mating, then take up seminal fluid antigens and traffic from the uterus to the dLN.34 To investigate whether altered DC dynamics or phenotype might contribute to the altered T cell response, CD11c+ DCs were quantified by immunohistochemistry (IHC) in the uterine endometrium at estrus and on d0.5 pc (Fig. 6a). There were more DCs in the endometrium of miR-155−/− mice than in miR-155+/+ mice at estrus, but unlike miR-155+/+ mice where CD11c+ DCs infiltrated the stroma subjacent to the luminal epithelium on d0.5 pc, there was no mating-associated DC increase in miR-155−/− mice (Fig. 6b).
CD11c+ dendritic cells were detected by IHC in uterus or by flow cytometry in uterus and dLN recovered from miR-155+/+ (+/+) and miR-155−/− (−/−) mice at estrus (est) or after mating (d0.5 pc and d3.5 pc). Representative images of CD11c+ dendritic cells detected in uterus by IHC at estrus and d0.5 pc are shown (a; scale bar = 50 µM). CD11c+ dendritic cells were quantified using ImageJ and expressed as percentage of positivity/area of endometrial tissue. Additional data are flow cytometric analysis of the effects of mating and genotype on dendritic phenotype in the uterus on d0.5 pc (c–e), and dendritic cell number in the uterus and dLN on d3.5 pc (f, g). Panels are the proportion of CCR7+CD11c+F4/80− dendritic cells in the uterus at estrus and d0.5 pc (c); the MFI of CCR7 within CD11c+F4/80− dendritic cells in the uterus at estrus and d0.5 pc (d); representative histogram of CCR7 expression in uterine dendritic cells at estrus (e), number of CD11c+F4/80− dendritic cells in the uterus at estrus and d3.5 pc (f), and number of CD11c+F4/80− dendritic cells in the dLN at estrus and d3.5 pc (f). Symbols depicting values from individual mice (n = 7–9/group) and mean ± SEM are shown. Differences between groups were assessed by one-way ANOVA with Sidak t test (*p < 0.05 vs. same time point in miR-155+/+, #p < 0.05 vs. estrus within genotype). The overall effect of genotype and mating was assessed by two-way ANOVA (∧p < 0.05 difference attributable to genotype; τp < 0.05 difference attributable to mating). Additional data on DC number and phenotype in the uterus, dLN, and spleen are provided in Supplementary Figs. S5–S7.
Flow cytometry revealed additional changes in uterine DCs on d0.5 pc. Notably a smaller proportion of uterine CD11c+F4/80− DCs expressed the chemokine receptor CCR7 in estrus mice (Fig. 6c–e), but major histocompatibility complex II (MHCII), CD80, or CD86 expression were not consistently different (Supplementary Fig. S5). The change in CCR7 was mirrored in the spleen DCs of miR-155−/− mice (Supplementary Fig. S6).
To investigate whether changes in uterine DCs were sustained to implantation and reflected in the dLN, CD11c+F4/80− DCs in the uterus and dLN were analyzed on d3.5 pc. Uterine DCs remained higher than at estrus in miR-155+/+ but not in miR-155−/− mice (Fig. 6f). By this time, dLN DCs were increased 2.2-fold in miR-155+/+ mice, presumably due to immigration of uterine DCs, but this did not occur in miR-155−/− mice, where there were 76% fewer DCs on d3.5 pc than in wild-type controls (Fig. 6g). The phenotype of uterine DCs and dLN DCs on d3.5 pc, as measured by MHCII, CD80, and CD86 expression, was comparable in miR-155+/+ and miR-155−/− mice (Supplementary Fig. S7).
miR-155 deficiency alters macrophages in the uterus and dLN
Macrophages can also modulate the adaptive immune response of early pregnancy.34,61 IHC of F4/80+ macrophages on d0.5 pc showed comparable numbers in the uterine endometrial stroma at estrus and an increase after mating in both miR-155+/+ and miR-155−/− mice (Fig. 7a, b).
F4/80+ macrophages were detected by IHC in uterus, or by flow cytometry in uterus and dLN recovered from miR-155+/+ (+/+) and miR-155−/− (−/−) mice at estrus (est) or after mating (d0.5 pc and d3.5 pc). Representative images of F4/80+ macrophages detected in uterus by IHC at estrus and on d0.5 pc are shown (a; scale bar = 50 µM). F4/80+ macrophages were quantified using ImageJ and expressed as percentage of positivity/area of endometrial tissue. Additional data are flow cytometric analysis of the effects of mating and genotype on macrophage phenotype in the uterus (c–e), and macrophage number in the uterus and dLN on d3.5 pc (f, g). Panels are the proportion of CD86+CD11b+F4/80+ macrophages in the uterus at estrus and d0.5 pc (c), the MFI of CD86 within CD11b+F4/80+ macrophages in the uterus at estrus and d0.5 pc (d), representative histogram of CD86 expression in uterine macrophages on d0.5 pc (e), number of CD11b+F4/80+ macrophages in the uterus at estrus and d3.5 pc (f), and the number of CD11b+F4/80+ macrophages in the dLN at estrus and d3.5 pc (g). Symbols depicting values from individual mice (n = 7–9/group) and mean ± SEM are shown. Differences between groups were assessed by one-way ANOVA with Sidak t test (*p < 0.05 vs. same time point in miR-155+/+, #p < 0.05 vs. estrus within genotype). The overall effect of genotype and mating was assessed by two-way ANOVA (∧p < 0.05 difference attributable to genotype; τp < 0.05 difference attributable to mating). Additional data on macrophage number and phenotype in the uterus, dLN and spleen are provided in Supplementary Figs. S8–S10.
Flow cytometry showed no change in the proportion of CD86+ macrophages among CD11b+F4/80+ uterine macrophages in miR-155−/− mice, but CD86 expression measured as MFI was substantially reduced, particularly after mating on d0.5 pc (Fig. 7c–e). Uterine macrophages also exhibited a small increase in CD80, but MHCII and CCR7 were unchanged (Supplementary Fig S8). Spleen macrophages showed a similar reduction in CD86 (Supplementary Fig S9).
By d3.5 pc, uterine macrophages had returned to estrus numbers regardless of miR-155 status (Fig. 7f). In contrast, the number of macrophages in dLN of miR-155+/+ mice was increased by 63%, but no increase was evident in miR-155−/− mice where macrophage numbers were 39% fewer than in miR-155+/+ females (Fig. 7g). There was no difference in uterine or dLN macrophage CD80, CD86, and MHCII expression on d3.5 pc due to miR-155 deficiency (Fig. S10).
miR-155 deficiency elicits reduced Csf2 and expression in the uterine immune response to seminal fluid
The recruitment of macrophages and DCs into the uterus, and their exodus to the dLN is regulated by chemokines induced in the endometrium during the inflammatory response to seminal fluid. Quantitative polymerase chain reaction (qPCR) analysis showed strong induction of Cxcl10 and CCR7 ligand Ccl19 after mating in miR-155+/+ mice but not in miR-155−/− mice (Fig. 8a, b). A second CCR7 ligand Ccl21 was also reduced in the endometrium of miR-155−/− mice after mating (Fig. 8c). Other DC-regulating cytokines Csf2, Csf3, Tnf, Il6, Ccl2, Cxcl1, and Cxcl2 were robustly induced in miR-155+/+ mice, but Tnf, Il6, Ccl2, Cxcl1, and Cxcl2 appeared aberrantly regulated in a proportion of estrus miR-155-deficient mice, such that significant induction after mating was not evident (Supplementary Fig. S11).
Cytokine expression was quantified by qPCR in the uterus recovered from miR-155+/+ (+/+) and miR-155−/− (−/−) mice at estrus (est) or after mating (8 h pc). Data are expression of Cxcl10 (a), Ccl19 (b), and Ccl21 (c) relative to Actb reference. Symbols depict values from individual mice (n = 7–10/group) and mean ± SEM values are shown. Effects of mating and genotype were assessed by one-way ANOVA with Sidak t test (*p < 0.05 vs. same time point in miR-155+/+, #p < 0.05 vs. estrus within genotype). The overall effect of genotype and mating was assessed by two-way ANOVA (^p < 0.05 difference attributable to genotype; τp < 0.05 difference attributable to mating).
Maternal miR-155 deficiency elevates lipopolysaccharide (LPS)-induced fetal loss
Impaired Tregs can compromise capacity to withstand inflammatory challenge in later gestation.62,63 To investigate the contribution of miR-155 to sustaining robust tolerance in pregnancy, miR-155+/+ and miR-155−/− mice were mated with BALB/c males and administered LPS or phosphate-buffered saline (PBS) on d9.5 pc. A low dose of LPS, just below the threshold dose required to induce inflammation-mediated fetal loss in wild-type mice, was chosen based on previous studies.64 At autopsy just prior to term on d17.5 pc, only 58% of LPS-treated miR-155−/− mice remained pregnant with at least one viable fetus, compared with 100% of miR-155+/+ mice (Fig. 9a). Fetal resorption was 59% in miR-155−/− compared with 12% in miR-155+/+ mice (Fig. 9b, g), yielding 60% fewer viable fetuses (Fig. 9c, g). After LPS administration, fetal weight was reduced by 8.3% in miR-155−/− mice compared to miR-155+/+ mice (Fig. 9d). Although placental weight was unchanged (Fig. 9e), there was a greater LPS-induced reduction in the fetal:placental weight ratio (a measure of placental function) in miR-155−/− mice after LPS exposure (Fig. 9f). There was no impact of miR-155 deficiency on fertility or fetal outcomes in mice given PBS.
miR-155+/+ (+/+) and miR-155−/− (−/−) females (n = 14–21/group) were mated and administered LPS or vehicle (PBS) on d9.5 pc and autopsied on d17.5 pc. The proportion of mated females with at least 1 viable implant (a), the proportion of implantation sites per pregnant female undergoing resorption (b), the number of viable implantation sites per mated mouse (c), fetal weight (d), placental weight (e), fetal:placental weight ratio (f), and representative macroscopic images of uterine horns following treatment with arrows indicating fetal resorption (g) are shown, with symbols depicting values from individual mice with mean ± SEM values (b, c) or estimated marginal mean ± SEM (d–f). The effect of genotype and mating was assessed by Chi-square analysis (a), by non-parametric Kruskal–Wallis test with post hoc Dunn’s multiple comparison test (b, c), and by mixed model analysis using the mother as subject and litter size as co-variate (d–f) (*p < 0.05 vs. same treatment in miR-155+/+; #p < 0.05 vs. PBS within genotype), or by two-way ANOVA (a–f) (∧p < 0.05 difference attributable to genotype; τp < 0.05 difference attributable to mating).
Transfer of miR-155+/+ Treg cells protects against inflammation-induced fetal loss
To determine whether wild-type miR-155+/+ Treg cells can rescue the pregnancy deficit in miR-155−/− mice, Treg cells pooled from the lymph nodes and spleen of pregnant miR-155+/+ mice were administered to miR-155−/− mice on d8.5 pc before LPS administration on d9.5 pc. Treg transfer reduced fetal resorption by 87% compared to controls given CD4+CD25− Tconv cells and by 85% compared to mice given vehicle control (Fig. 10a, b), with a corresponding trend toward more viable implantation sites (Fig. 10c). Treg treatment did not alter fetal or placental weights or the fetal:placental weight ratio (data not shown).
CD4+CD25+ Treg cells (Treg) and CD4+CD25− T cells (Tconv) isolated from the lymph nodes and spleen of pregnant miR-155+/+ mice were administered to recipient miR-155−/− mice (2–4 × 105 Treg or Tconv cells, or Veh, on d8.5 pc). Mice then received LPS or vehicle (PBS with 2% BSA) on d9.5 pc and pregnancy parameters were assessed on d17.5 pc. The proportion of mated females with at least one viable implant (a), the proportion of implantation sites per pregnant female undergoing resorption (b), and the number of viable implantation sites per mated mouse (c) are shown, with symbols depicting values from individual mice with mean ± SEM values (b, c). The effect of genotype and mating was assessed by Chi-square analysis (a) and by non-parametric Kruskal–Wallis test with post hoc Dunn’s multiple comparison test (b, c). (∧p = 0.068 vs. Veh or Tconv).
Discussion
A sufficient population of functionally competent Treg cells must be present in the uterus and its dLNs to mediate maternal immune tolerance of embryo implantation.5,14 Tregs recruited into the endometrial lining of the uterus engage with a network of DCs, macrophages, and uterine natural killer cells to facilitate decidual transformation of the endometrium and support placental morphogenesis and fetal growth.2 Deficiency in Treg cells at implantation causes unresolved inflammatory activation and uterine vascular aberrations that ultimately dispose toward later miscarriage, preeclampsia, and hypertensive disorders of gestation.16,25,26,27 MicroRNAs are likely to be important regulators of Treg cell generation and function in pregnancy tolerance, and several are differentially expressed in clinical conditions of pregnancy, but their specific contributions are not yet defined.
miR-155 is an ancient and highly conserved microRNA (miRNA) present in all jawed vertebrates studied to date.65,66 Its evolution coincided with emergence of an adaptive immune system implying a highly conserved role across species, through potent regulation of myeloid and lymphoid cell behavior.66 Here we show that miR-155 is induced in the uterus and uterus-dLNs in early pregnancy. We find that the normal surge in Treg cell activation and expansion induced by seminal fluid contact is dysregulated in the absence of miR-155. The stunted Treg response imparts susceptibility to a “second-hit” inflammatory challenge, such that miR-155-deficient mice exhibit fivefold higher fetal loss in response to low-dose LPS in mid-gestation, which can be rescued by administration of wild-type Treg cells.
The dLN is the primary site where Treg cells proliferate after conception to provide a circulating pool for recruitment into the uterus.18 Activation and proliferation of CD4+ T cells commences in the dLN in the hours following coitus, after DCs recruited into the endometrial stroma take up male alloantigens delivered in seminal fluid and then traffic to the dLN via the afferent lymphatics.31,34 The presence of abundant TGFβ and other immune-regulating agents in seminal fluid ensures the resulting T cell response is skewed toward Treg cells and against T helper type 1 (Th1) and Th17 cell generation.17,31 This antigen-driven proliferation of Treg cells within the dLN builds on a hormone-dependent increase in circulating Treg cells elicited by the estrogen surge accompanying ovulation.28 It involves both pTreg and tTreg proliferation and functional maturation, including epigenetic modulation of tTreg cells by demethylation of CpG sites in the CNS1 enhancer region of the FOXP3 promoter.19
Expression of miR-155 was upregulated in both the uterus and the dLN, from 36 h after mating through to initiation of embryo implantation around 48 h later on d3.5 pc. A minor elevation in miR-155 occurred after ovulation in unmated mice, consistent with either estrogen-regulated recruitment of immune cells expressing miR-155 or estrogen-controlled elevated expression of miR-155, as reported for human breast cancer cells.67 However, the increase attributable to hormones was at least a scale of magnitude lower than the increase after mating. Induction of miR-155 in mated mice required contact with both sperm and seminal plasma fractions of seminal fluid from an allogeneic male, since mating with syngeneic males, or males from which seminal fluid components were removed by surgical excision of the seminal vesicle gland, did not induce miR-155 beyond the small estrus cycle-associated increase. miR-155 deficiency curtailed the CD4+ T cell response, with a disproportionate impact on Treg cells, causing substantial reduction in the number of dLN Treg cells on d3.5 pc of early pregnancy, when antigen-driven T cell proliferation normally peaks.34,58 As a consequence, the Treg cell pool and the Treg:Teff ratio were substantially reduced for the course of pregnancy. These findings are consistent with involvement of this miRNA in the events driving T cell proliferation that commence in the lymph node shortly following mating and fit with studies in mice showing that maximum expansion of the uterine Treg pool requires alloantigens.68
Reduced Treg cells in the absence of miR-155 appeared to arise as the consequence of both reduced size of the Treg cell pool before conception and impaired proliferation of CD4+ T cells following seminal fluid contact. miR-155-deficient mice have substantially smaller Treg cell populations in the spleen and peripheral lymph nodes in steady-state conditions.43,44,69 Indeed we found that Treg cells comprised approximately two-fold smaller proportions of the total CD4+ T cell population in the dLN, spleen, and MLN of non-pregnant miR-155-deficient mice. However, this does not explain the four-fold fewer Treg cells in early pregnancy, implicating miR-155 in additional events regulating the periconception immune response.
Both pTreg and tTreg cells are implicated in mediating maternal immune tolerance in pregnancy.19,20,21 Here we found that the NRP1+ thymic and NRP1− peripheral Treg cell compartments are both reduced by miR-155 deficiency. NRP1 is generally viewed as a useful marker to distinguish pTreg and tTreg cells.6,7 There are concerns that in some hyper-inflammatory conditions NRP1 can be induced on pTreg cells.70 However, our data show that, even before mating, NRP1+ Treg cells are prevalent in the uterus and dLN, so are highly likely to be of thymic origin.19 A smaller tTreg pool has been attributed to reduced thymic output of Treg cells associated with impaired SOCS1 control of IL-2 signaling, upon which Treg cells are notably dependent for their generation and homeostasis.41,44 In addition, thymus-derived Treg require miR-155 to mediate regulatory signaling activated by the C-type lectin CD69.69 Our conventional and tSNE analysis clearly showed the effects of miR-155 deficiency on both populations before and during pregnancy, consistent with an impact of miR-155 on thymic generation of naive CD4+ T cells as well as tTregs.36
Within mated females, miR-155 deficiency caused a modest reduction in the intensity of CTLA4 expression in Treg cells. Since CTLA4 is a negative regulator of Treg cell homeostasis that acts as a “brake” on Treg cell proliferation,71 reduced CTLA4 could exert positive pressure on Treg cell proliferation, perhaps explaining the relatively normal Ki67 expression in miR-155-deficient Treg cells. CTLA4 is also a mediator of suppressive competence in Treg cells.72 However, extensive previous studies consistently report no change in suppressive competence of Treg cells from miR-155−/− mice.44,45 Moreover, miR-155-deficient mice do not exhibit autoimmunity and are resistant to experimental autoimmune encephalomyelitis,73 and miR-155-deficient Treg cells maintain tolerance after transfer to miR-155-replete hosts.44 These observations imply that miR-155 predominantly contributes to Treg cell differentiation and proliferation but does not affect suppressive function44 and suggest redundancy in the role of miR-155 to reinforce the FOXP3 transcriptional program.74
Treg cells, particularly effector/memory Treg cells, express higher levels of miR-155 than other CD4+ T cells.43,75 Given the requirement for alloantigen and timing of miR-155 increase in the uterus after mating, which coincides with alloantigen-dependent Treg cell activation, proliferation, and recruitment,18 we infer that Treg cells are the major cellular source of miR-155 in the uterus. However, activated DCs and macrophages also express miR-155 and their phenotypes and antigen-presenting function are modulated by this miRNA38,39,76 through mechanisms that include miR-155-mediated suppression of c-Fos and Arg-2 to upregulate MHCII and costimulatory molecules CD80, CD86, and CD40.48,49
Consistent effects of miR-155 deficiency on DC co-stimulatory molecule expression were not evident in the current study. However, there were substantially fewer DCs in the dLN accompanied by greater DC numbers in the uterus, indicating impaired DC exodus from the uterus and trafficking to the dLN. DCs from miR-155-deficient mice exhibited lower expression of CCR7, a chemokine receptor controlling DC lymph node homing in homeostatic and inflammatory conditions.77,78 A previous study showed that miR-155 deficiency causes altered DC emigration to dLN in a murine tumor model.49 DCs from mammary tumors had reduced expression of CCR7 due to miR-155 suppression of Jarid2, causing reduced DNA methylation at the Ccr7 locus.42,49 A similar induction of CCR7 by miR-155 occurs in human peripheral blood monocytes (PBMCs).79
Macrophages are recruited into the uterus after mating and exhibit features indicating competence to regulate T cell activation.80 Previous studies demonstrate that miR-155 induction occurs after macrophage activation and influences their differentiation program.46 In the absence of miR-155, fewer macrophages were present in the dLN on d3.5 pc. While uterine macrophages showed an altered phenotype with lower CD86, whether and how this is linked to reduced dLN macrophages is unclear. There is evidence that inflammatory monocytes recruited into the uterus in early pregnancy acquire a regulatory phenotype or perhaps differentiate into myeloid DCs.81 Thus fewer macrophages in the absence of miR-155 might reinforce DC deficiency and contribute to the impaired Treg cell response.
Altered uterine transcription of chemokines controlling DC and macrophage migration and phenotype implies indirect as well as cell-intrinsic effects of miR-155 on uterine myeloid cells. Notably, miR-155 deficiency caused reduced expression of the CCR7 ligands Ccl19 and Ccl21. CCL19 and CCL21 act at multiple levels to control DC recruitment into, and exodus from, epithelial tissues.78 Langerhan’s cells and dermal DCs require CCL21 signaling via CCR7 to emigrate through the dermis to the afferent lymphatics, after antigen uptake in the skin.82 CCL19 is rapidly induced in the uterine epithelium by seminal fluid factors after mating in mice.18 Mice with null mutation in Ccl21 or Ccr7 exhibit deficient recirculation of epithelial DCs to lymph nodes under homeostatic conditions,77,83 with dysregulated peripheral tolerance through effects on T cell immigration and priming, as demonstrated in the respiratory and gastrointestinal tracts.84,85
We found that miR-155 deficiency also impaired the expected post-mating induction of interferon-induced chemokine Cxcl10 (IP-10), a product of myeloid-derived DCs, that modulates their capacity to attract and stimulate effector T cells.86 miR-155-deficient mice similarly have reduced Cxcl10 expression in brain tissue after traumatic injury.87 Tnf, Il6, Ccl2, Cxcl1, and Cxcl2 also showed differential expression, with greater variability particularly in estrus mice. This could reflect miR-155 regulation of cytokine synthesis by myeloid leukocytes and/or uterine epithelial cells, cell lineages that both contribute substantially to uterine cytokine synthesis,88 and exhibit altered inflammatory cytokine production in epithelia of miR-155 null mutant mice.46,89 Clear evidence of epithelial cell-intrinsic regulation of cytokine synthesis by miR-155 comes from studies of lung epithelium in miR-155 null mutant mice89 and findings that TGFβ induces miR-155 through Smad4 in mammary gland epithelium.90 miR-155 attenuates cytokine gene expression through targets in the nuclear factor-κB pathway through targeting SOCS147 and JNK46 in leukocytes and RhoA90 and PKIα89 in epithelial cells, as well as other transcriptional activators and epigenetic regulators.42,89 Macrophage regulation of epithelial cell phenotype in the uterus affords additional complexity in the regulatory networks potentially impacted by miR-155.91 The precise contributions of epithelial and myeloid lineages to uterine miR-155 expression, and the molecular mechanisms by which miR-155 controls cytokine production in these uterine cell lineages, remains to be determined.
Despite their altered Treg cell population, allogeneic pregnancy progressed normally in unchallenged miR-155-deficient mice. This could reflect that, within the Teff compartment, miR-155 deficiency causes a shift toward Th2 cells, not Th1 cells that are detrimental to pregnancy.36,39 However, there was a profound adverse impact of miR-155 deficiency in maternal capacity to withstand a “second-hit” of low-dose LPS to induce a systemic inflammatory response in mid-gestation, causing extensive fetal loss and fetal growth restriction. Several previous studies demonstrate that LPS elicits placental injury and fetal loss through induction of IL1b, Il6, Tnf, and other pro-inflammatory genes in gestational tissues,62,64 and sensitivity to fetal loss is impacted by availability of competent Treg cells.62,63 The reproductive data reported herein is from pregnancies sired by wild-type males yielding fetuses that are haplosufficient for miR-155, consistent with a requirement for maternal as opposed to fetal miR-155 in deflecting inflammatory challenge. Fetal miR-155 is not essential for development as we observe no overt detrimental effect in matings between homozygous null mutant male and female mice (data not shown).
Fetal loss was attributable to Treg inability to modulate and resolve inflammation, since transfer of miR-155-sufficient Treg cells rescued the phenotype. Treg cells are well known to exert potent anti-inflammatory activity, acting in many infection-associated and sterile injury settings to suppress and resolve inflammation.92 A similar failure by incompetent Treg cells to control mid-gestation inflammation is seen in IL-10-deficient mice, which exhibit Treg cell instability in pregnancy, accompanied by elevated susceptibility to inflammation-induced fetal loss.62 This is consistent with a limiting function of Treg cell number or stability that is not evident under steady-state conditions but manifests when infectious agents or environmental factors elicit an elevated inflammatory load.
Defining the critical regulatory networks controlling Treg cells in pregnancy is important in order to tackle human gestational disorders where impaired Treg cell numbers and/or phenotype are implicated in the underlying placental insufficiency and inflammatory pathology.5 There is compelling evidence linking insufficient Treg cells in the uterine decidua of early pregnancy with implantation failure, recurrent miscarriage, and preeclampsia.2,4 Failure of the normal expansion of Helios+ tTreg and pTreg during early pregnancy is linked with these conditions, and recent data implicate paternal alloantigens in driving the expansion of decidual Tregs93 and suggest clonal expansion of Helios+ tTreg and pTreg fails in infertile women,4 pointing to a key role for the periconception and early pregnancy environment. It is possible that insufficient miR-155 is a factor contributing to disrupted Treg cell responses in women, since altered miR-155 expression is reported in pregnancy disorders. In recurrent miscarriage, miR-155 is one of the five miRNAs identified as reduced in PBMCs in the first trimester compared with healthy controls.56 In women experiencing infertility associated with repeated implantation failure, miR-155 is downregulated in peripheral blood T cells compared to fertile women.55 In women with preeclampsia and a small-for-gestational-age fetus, miR-155 was one of the seven miRNAs upregulated in the placenta57 raising the question of whether sustained expression beyond implantation is counterproductive. The findings of the current study suggest that miR-155 deficiency might be part of a causal mechanism underlying the deficiencies in Tregs that have been well described in women with these common infertility and pregnancy disorders.2,4
In summary, the current study expands current knowledge to show that miR-155 is required for normal pregnancy tolerance, through pivotal roles in both the innate and adaptive immune compartments. In particular, miR-155 deficiency constrains the normal expansion in the CD4+ Treg cell pool driven by seminal fluid contact at conception, as a consequence of both reduced Treg cells in dLN before pregnancy, as well as compromised capacity for DCs to stimulate further expansion in response to paternal alloantigens. Although implantation and early fetal development progress normally, absence of miR-155 incapacitates the Treg cell pool leaving the placenta and fetus more vulnerable to inflammatory injury. These observations indicate that evaluating the link between miR-155 and pregnancy tolerance in women is warranted, since altered miR-155 and insufficient Treg cells are implicated in common pregnancy disorders.
Materials and methods
Mice
Experimental protocols were approved by the University of Adelaide Animal Ethics Committee (approval number: M-2014-023, M-2017-006). Bic/miR-155 (miR-155−/−) mice36 were sourced from The Jackson Laboratory (Bar Harbor, ME, USA) and bred in-house at University of Adelaide. C57Bl/6J (B6; miR-155+/+) wild-type control mice were from Animal Resource Centre, Perth. BALB/c male mice were from Laboratory Animal Services, University of Adelaide. Vasectomized (vas), seminal vesicle-excised (svx), and seminal vesicle-excised/vasectomized (svx/vas) BALB/c males were prepared as previously described.30,32 All mice were co-housed in specific pathogen-free conditions on a 12-h light–dark cycle with food and water ad libitum. Experimental females were 8–16 weeks and BALB/c stud males were aged 3–12 months. Estrous cycle stage was determined from vaginal lavage cells. For experimental matings, one to three 8–12-week-old B6 females were caged with intact, vas, svx, or svx/vas stud males. Females mated with intact or vas males were checked for the presence of vaginal plugs, whilst mating with svx males and svx/vas males was confirmed by video recording of coitus.30 The morning after mating was designated d0.5 pc.
RNA preparation and qPCR
miR-155+/+ and miR-155−/− females were killed at various stages of the estrous cycle or at specific time points (8, 16, 36, 60, 84 h) following mating. dLN or the endometrial layer of the uterus were collected as described30 and frozen in liquid N2. Tissue was homogenized (PowerLyzer 24, Qiagen) and RNA extracted using the miRNeasy Extraction Kit (Qiagen) and then treated with TURBO DNA-free™ (Thermo Fisher, Waltham, MA) following the manufacturer’s instructions. Absence of genomic DNA contamination was confirmed as described.94 RNA was quantified (Nano-Drop Spectrophotometer ND-1000, Thermo Fisher) and RNA integrity was analyzed by Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) to confirm RNA integrity number > 7.
For TaqMan assays, mmu-miR-155 (assay ID, 002571, Thermo Fisher) and U6snRNA (assay ID, 001973, Thermo Fisher) were reverse transcribed from 10 ng RNA using the TaqMan miRNA Reverse Transcription Kits (Thermo Fisher) using the manufacturer’s instructions. miRNA (0.7 ng) was amplified using 1× Taqman Universal PCR master mix II, no UNG (Thermo Fisher), and 1× Taqman miRNA PCR primer probes in a QuantStudio 3 Real-Time PCR System (Thermo Fisher) at 50 °C for 2 min, 95 °C for 10 min, then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The delta C(t) method95 was used to calculate miR-155 abundance normalized to U6snRNA reference.
For cytokine gene expression analysis, cDNA was reverse transcribed from 1000 ng total cellular RNA primed with 50 ng random hexamers employing a Superscript-IV Reverse Transcriptase Kit (Life Technologies) according to the manufacturer’s instructions and stored at −20 °C. Primer pairs (Supplementary Table 1) were optimized and validated as described.30 PCR amplification was performed on 20 ng of cDNA with PCR primers (details in Supplementary Table 1) and 1× PowerUp SYBR Green PCR master mix (Life Technologies) using a QuantStudio 3 Real-Time PCR System Sequence Detection System (Thermo Fisher) and the following conditions: 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles of 95 °C for 1 s and 60 °C for 30 s. The delta C(t) method95 was used to calculate mRNA abundance normalized to Actb reference.
Flow cytometry
Single-cell suspensions were prepared from the dLN, spleen, and MLN of miR-155+/+ and miR-155−/− females in estrus or d3.5, 9.5, or 15.5 pc. Whole uterus was minced using fine scissors and digested at 37 °C in 1 mg/ml Type I collagenase, 2.5 mg/ml DNase (Sigma-Aldrich, St Louis) in RPMI-1640 with 10% fetal calf serum (FCS), and 0.015 M HEPES. Cells were washed twice in fluorescent-activated cell sorting (FACS) buffer (0.1% bovine serum albumin (BSA), 0.5% sodium azide in PBS), filtered by 70-µm cell strainer, then incubated with fixable viability stain (FVS) 620 (1:1000 in PBS, BD Biosciences, San Jose, CA), washed in FACS buffer, and incubated with Fc receptor block (BD Biosciences). Cells were then stained with fluorophore-conjugated monoclonal antibodies (mAbs) CD4•APC-Cy7 (1:100), CD25•PE-Cy7 (1:100; both BD Biosciences), and NRP1•BV421 (1:75; Biolegend, San Diego, CA) in FACS buffer for 30 min at 4 °C. Labeled cells were washed in FACS buffer, fixed, and permeabilized with Intracellular Fixation and Permeabilization Buffer (Thermo Fisher) using the manufacturer’s instructions, then labeled with antibodies Ki67•FITC (SolA15, 1:125; Thermo Fisher), FOXP3•APC (FJK-16s, 1:100; Thermo Fisher), and CTLA4•PE (1:100; BD Biosciences) and analyzed using a FACS Canto II flow cytometer with the FACSDiva software (BD Biosciences). For quantification of absolute T cell subset numbers, count beads (CountBright™ Count Beads, Thermo Fisher) were added to a 10% aliquot of cells labeled with FVS620 and CD4•APC-Cy7.
For phenotype samples, gates were established to include viable lymphocytes and exclude debris and doublets using forward scatter (FSC) and side scatter profiles and the viability dye. Lymphocytes positive for CD4 were gated (Fig. S12), and CD25+FOXP3+CD4+ cells were defined as Treg cells. Gates were established within the CD4+CD25+FOXP3+ Treg cell population to distinguish NRP1− (pTreg) and NRP1+ (tTreg) cells. Ki67 and CTLA4 were measured within total Treg, tTreg, and pTreg populations. The MFI for FOXP3 and CTLA4 were calculated on the FOXP3+ and CTLA4+ populations, respectively.
For count bead samples, count beads were gated based on fluorescent signal and scatter profiles. The lymphocyte population was established using the scatter profiles excluding debris and displayed as a CD4 vs. viability dye dot plot to gate viable CD4+ cells. The ratio of count beads to CD4+ cells was used to calculate the total number of the cell types of interest, as previously.18 In all samples, gates were established using unlabeled, fluorescence minus one and isotype-matched negative controls.
DCs and macrophages were incubated with FVS620 and Fc receptor block, then fluorophore-conjugated mAbs CD11B•APC-Cy7 (M1/70, 1:100), CD11C•BV510 (1:100), F4/80•PE (T45-2342, 1:50), MHCII•Alexa Fluor® 488 (M5/144.15.2, 1:100; all BD Biosciences), CD80•PE-Cy7 (16-10A1, 1:100), and CD86•APC (GL1, 1:100; both Thermo Fisher) for 15 min at room temperature. Labeled cells were then washed in FACS buffer and resuspended in PBS prior to FACS analysis. In some experiments, cells from the LN, spleen, and uterus were rested in 200 µl Iscove’s Modified Dulbecco’s Media (Thermo Fisher) + 10% FCS for 2 h under cell culture conditions to recover CCR7. Cells were then incubated with FVS620 and Fc receptor block, and mAb against CCR7•BV605 (1:100) at 37 °C for 15 min. An mAb cocktail of CD45•BUV395 (1:100), CD11B•APC-Cy7 (1:100), CD11C•BB515 (1:100), F4/80•BV421 (1:100), MHCII•BV711 (1:100; all BD Biosciences), CD80•PE-Cy7 (1:100), and CD86•APC (1:100; both Thermo Fisher) was added for 30 min at room temperature in Brilliant Stain Buffer Plus (BD Biosciences, as per the manufacturer’s instructions). Labeled cells were then washed in FACS buffer and resuspended in PBS prior to FACS analysis.
DC and macrophage phenotypes were analyzed as above. Gates were established to include viable leukocytes and exclude debris and doublets using FSC and side scatter profiles and the viability dye (Supplementary Fig. S12). CD45+CD11B+F4/80+ cells were defined as macrophages, and CD45+CD11C+F4/80− cells as DCs. Gates were established within macrophage and DC populations to measure MHCII, CD80, and CD86.
tSNE analysis
Single cells from dLN of estrus and d3.5 pc mated miR-155+/+ and miR-155−/− mice labeled with antibodies CD4-APC•Cy7, CD25•PE-Cy7, Ki67•FITC, NRP1•BV421, CTLA4•PE, and FOXP3•APC were analyzed by flow cytometry as above, prior to processing for tSNE analysis as described,19,96 gated to include only CD4+ T cells, using a perplexity of 70, and a Barns–Hut approximation value of 0.5. Once the algorithm was complete, the tSNE approximation was then applied to all events and manually clustered with the aid of density and contour plots. The proportion and MFI of each cluster were derived from the FCS Express software. A Z-score heat map of MFI for phenotypic markers was generated using Heatmapper software.97
Immunohistochemistry
Whole uterus from miR-155+/+ and miR-155−/− mice was collected at estrus or d0.5 pc after mating with BALB/c males, trimmed of fat and connective tissue, embedded in Tissue-Tek OCT compound (Sakura Finetek, Torrance, USA), and stored at −80 °C. Sections (v) were cut using a Leica CM 1850 cryostat (Leica Microsystems, NSW, Australia), fixed onto Colourfrost microscope slides (Trajan, VIC, Australia), and stored with desiccant at −80 °C until staining.
After thawing, sections were fixed in 96% ethanol for 10 min, washed in PBS, and blocked in 1% BSA in PBS (PBS-BSA). To detect macrophages, sections were incubated with 15% rabbit serum in PBS-BSA for 30 min, then rat anti-mouse F4/80 (Agilent Technologies; 1:400 in 1.5% rabbit serum, PBS-BSA) overnight at 4 °C, and then biotinylated rabbit anti-rat IgG (Agilent; 1:500 in 1.5% rabbit serum, PBS-BSA) for 40 min at room temperature. To detect DCs, sections were incubated with biotinylated rat anti-mouse CD11c (Biolegend; 1:50 in PBS-BSA) overnight at 4 °C. Antibody detection utilized the ABC HRP Kit (Vector Laboratories, Burlingame, CA) and DAB Peroxidase Substrate Kit (Agilent Technologies, Santa Clara, CA), and sections were counterstained with hematoxylin (Sigma-Aldrich). Negative controls were incubated with normal rabbit serum or isotype-matched rat mAb in place of primary antibody. DAB was applied to detect endogenous peroxidase+ eosinophils. Images were captured using a Nanozoomer-XR Digital Slide Scanner (Hamamatsu Photonics, Hamamatsu, Japan) at a zoom equivalent to a ×40 objective lens. F4/80+ and CD11c+ cells were quantified using ImageJ excluding luminal epithelium and glandular epithelium from the analysis. The relative density of F4/80+ and CD11c+ cells was determined as percentage of positivity = 100 × (area of F4/80 or CD11c staining − area endogenous peroxidase staining)/area total staining (DAB plus hematoxylin).
Pregnancy outcome analysis
miR-155+/+ and miR-155−/− females mated with BALB/c males and with >2.0 g weight gain between d0.5 pc and d9.5 pc were administered LPS (Escherichia coli; Sigma-Aldrich; 1.0 μg in 200 μl PBS with 0.1% BSA; intraperitoneal) as described64 or PBS, with 0.1% BSA control at 1100 h on d9.5 pc, then killed by cervical dislocation at 1100–1400 h on d17.5 pc. Intact uteri were excised and examined for total number and the number of viable and resorbing implantation sites. Each viable fetus and placenta was dissected and weighed.
Treg cell transfer
CD4+CD25+ Treg cells were isolated from the dLN, MLN, inguinal, and brachial lymph nodes and spleen from pregnant d10.5 to 13.5 pc miR-155+/+ mice using the EasySepTM Mouse CD4+CD25+ Regulatory T Cell Isolation Kit II (STEMCELL Technologies, Vancouver) according to the manufacturer’s instructions. Cells were eluted in PBS with 2% FCS and confirmed to be >90% viable CD4+CD25+ by flow cytometry using CD4•APC-Cy7 and CD25•PE-Cy7 as above. Recipient miR-155−/− mice were administered 2–4 × 105 CD4+CD25+ Treg or CD4+CD25− (Tconv) cells intravenously into tail vein on d8.5 pc. A control group were administered vehicle alone (PBS with 2% FCS). Mice were then given LPS or vehicle on d9.5 pc and assessed for late gestation pregnancy parameters on d17.5 pc as described above.
Statistics and data analysis
Data were assessed for normality using the D’Agostino and Pearson normality test. Flow cytometric data, viable litter size, and resorption rate was analyzed by two-way analysis of variance (ANOVA) to detect main effects of genotype and mating and one-way ANOVA with a post hoc Sidak test to detect differences between treatment groups. Data not normally distributed were transformed by Box–Cox transformation and analyzed as above. If transformation did not yield normal distribution, data were analyzed by non-parametric Kruskal–Wallis test with post hoc Dunn’s multiple comparisons test. Categorical pregnancy outcome data were compared by χ2 analysis. Fetal weight, placental weight, and fetal:placental weight ratio were analyzed by Mixed Model Linear Repeated-Measures ANOVA and post hoc least significant difference t test, with the mother as subject. Differences between groups were considered significant when p < 0.05.
References
Erlebacher, A. Mechanisms of T cell tolerance towards the allogeneic fetus. Nat. Rev. Immunol. 13, 23–33 (2013).
Robertson, S. A., Care, A. S. & Moldenhauer, L. M. Regulatory T cells in embryo implantation and the immune response to pregnancy. J. Clin. Investig. 128, 4224–4235 (2018).
Robertson, S. A. & Sharkey, D. J. Seminal fluid and fertility in women. Fertil. Steril. 106, 511–519 (2016).
Tsuda, S., Nakashima, A., Shima, T. & Saito, S. New paradigm in the role of regulatory T cells during pregnancy. Front. Immunol. 10, 573 (2019).
Guerin, L. R., Prins, J. R. & Robertson, S. A. Regulatory T-cells and immune tolerance in pregnancy: a new target for infertility treatment? Hum. Reprod. Update 15, 517–535 (2009).
Weiss, J. M. et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 209, 1723–1742 (2012).
Yadav, M. et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 209, 1713–1722, S1711–S1719 (2012).
Delgoffe, G. M. et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature 501, 252–256 (2013).
Chen, W. & Konkel, J. E. Development of thymic Foxp3(+) regulatory T cells: TGF-beta matters. Eur. J. Immunol. 45, 958–965 (2015).
Yamazaki, S. et al. Homeostasis of thymus-derived Foxp3+ regulatory T cells is controlled by ultraviolet B exposure in the skin. J. Immunol. 193, 5488–5497 (2014).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Panduro, M., Benoist, C. & Mathis, D. Tissue Tregs. Annu. Rev. Immunol. 34, 609–633 (2016).
Thornton, A. M. & Shevach, E. M. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 188, 287–296 (1998).
Aluvihare, V. R., Kallikourdis, M. & Betz, A. G. Regulatory T cells mediate maternal tolerance to the fetus. Nat. Immunol. 5, 266–271 (2004).
Shima, T. et al. Regulatory T cells are necessary for implantation and maintenance of early pregnancy but not late pregnancy in allogeneic mice. J. Reprod. Immunol. 85, 121–129 (2010).
Care, A. S. et al. Reduction in regulatory T cells in early pregnancy causes uterine artery dysfunction in mice. Hypertension 72, 177–187 (2018).
Robertson, S. A. et al. Seminal fluid drives expansion of the CD4+CD25+ T regulatory cell pool and induces tolerance to paternal alloantigens in mice. Biol. Reprod. 80, 1036–1045 (2009).
Guerin, L. R. et al. Seminal fluid regulates accumulation of FOXP3+ regulatory T cells in the preimplantation mouse uterus through expanding the FOXP3+ cell pool and CCL19-mediated recruitment. Biol. Reprod. 85, 397–408 (2011).
Moldenhauer, L. M. et al. Thymus-derived regulatory T cells exhibit Foxp3 epigenetic modification and phenotype attenuation after mating in mice. J. Immunol. 203, 647–657 (2019).
Samstein, R. M., Josefowicz, S. Z., Arvey, A., Treuting, P. M. & Rudensky, A. Y. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 150, 29–38 (2012).
Rowe, J. H., Ertelt, J. M., Xin, L. & Way, S. S. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature 490, 102–106 (2012).
Somerset, D. A., Zheng, Y., Kilby, M. D., Sansom, D. M. & Drayson, M. T. Normal human pregnancy is associated with an elevation in the immune suppressive CD25(+) CD4(+) regulatory T-cell subset. Immunology 112, 38–43 (2004).
Tilburgs, T. et al. Evidence for a selective migration of fetus-specific CD4+CD25bright regulatory T cells from the peripheral blood to the decidua in human pregnancy. J. Immunol. 180, 5737–5745 (2008).
Sasaki, Y. et al. Decidual and peripheral blood CD4+CD25+ regulatory T cells in early pregnancy subjects and spontaneous abortion cases. Mol. Hum. Reprod. 10, 347–353 (2004).
Jasper, M. J., Tremellen, K. P. & Robertson, S. A. Primary unexplained infertility is associated with reduced expression of the T-regulatory cell transcription factor Foxp3 in endometrial tissue. Mol. Hum. Reprod. 12, 301–308 (2006).
Winger, E. E. & Reed, J. L. Low circulating CD4(+) CD25(+) Foxp3(+) T regulatory cell levels predict miscarriage risk in newly pregnant women with a history of failure. Am. J. Reprod. Immunol. 66, 320–328 (2011).
Inada, K. et al. Characterization of regulatory T cells in decidua of miscarriage cases with abnormal or normal fetal chromosomal content. J. Reprod. Immunol. 97, 104–111 (2013).
Kallikourdis, M. & Betz, A. G. Periodic accumulation of regulatory T cells in the uterus: preparation for the implantation of a semi-allogeneic fetus? PLoS ONE 2, e382 (2007).
Robertson, S. A., Ingman, W. V., O’Leary, S., Sharkey, D. J. & Tremellen, K. P. Transforming growth factor beta-a mediator of immune deviation in seminal plasma. J. Reprod. Immunol. 57, 109–128 (2002).
Schjenken, J. E., Glynn, D. J., Sharkey, D. J. & Robertson, S. A. TLR4 signaling is a major mediator of the female tract response to seminal fluid in mice. Biol. Reprod. 93, 68 (2015).
Shima, T. et al. Paternal antigen-specific proliferating regulatory T cells are increased in uterine-draining lymph nodes just before implantation and in pregnant uterus just after implantation by seminal plasma-priming in allogeneic mouse pregnancy. J. Reprod. Immunol. 108, 72–82 (2015).
Robertson, S. A., Mau, V. J., Tremellen, K. P. & Seamark, R. F. Role of high molecular weight seminal vesicle proteins in eliciting the uterine inflammatory response to semen in mice. J. Reprod. Fertil. 107, 265–277 (1996).
McMaster, M. T., Newton, R. C., Dey, S. K. & Andrews, G. K. Activation and distribution of inflammatory cells in the mouse uterus during the preimplantation period. J. Immunol. 148, 1699–1705 (1992).
Moldenhauer, L. M. et al. Cross-presentation of male seminal fluid antigens elicits T cell activation to initiate the female immune response to pregnancy. J. Immunol. 182, 8080–8093 (2009).
Sharkey, D. J., Tremellen, K. P., Jasper, M. J., Gemzell-Danielsson, K. & Robertson, S. A. Seminal fluid induces leukocyte recruitment and cytokine and chemokine mRNA expression in the human cervix after coitus. J. Immunol. 188, 2445–2454 (2012).
Thai, T. H. et al. Regulation of the germinal center response by microRNA-155. Science 316, 604–608 (2007).
Grigoryev, Y. A. et al. MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes. J. Immunol. 187, 2233–2243 (2011).
O’Connell, R. M., Zhao, J. L. & Rao, D. S. MicroRNA function in myeloid biology. Blood 118, 2960–2969 (2011).
Rodriguez, A. et al. Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 (2007).
Turner, M. & Vigorito, E. Regulation of B- and T-cell differentiation by a single microRNA. Biochem Soc. Trans. 36(Pt 3), 531–533 (2008).
Yao, R. et al. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE 7, e46082 (2012).
Escobar, T. M. et al. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity 40, 865–879 (2014).
Kohlhaas, S. et al. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J. Immunol. 182, 2578–2582 (2009).
Lu, L. F. et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30, 80–91 (2009).
Stahl, H. F. et al. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PLoS ONE 4, e7158 (2009).
O’Connell, R. M., Taganov, K. D., Boldin, M. P., Cheng, G. & Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl Acad. Sci. USA 104, 1604–1609 (2007).
Wang, P. et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 185, 6226–6233 (2010).
Dunand-Sauthier, I. et al. Repression of arginase-2 expression in dendritic cells by microRNA-155 is critical for promoting T cell proliferation. J. Immunol. 193, 1690–1700 (2014).
Wang, J. et al. microRNA-155 deficiency impairs dendritic cell function in breast cancer. Oncoimmunology 5, e1232223 (2016).
Beyer, M. et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 12, 898–907 (2011).
Taganov, K. D., Boldin, M. P., Chang, K. J. & Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA 103, 12481–12486 (2006).
Zhou, H., Li, J., Gao, P., Wang, Q. & Zhang, J. miR-155: a novel target in allergic asthma. Int. J. Mol. Sci. 17, 1773 (2016).
Zhou, Q. et al. Decreased expression of miR-146a and miR-155 contributes to an abnormal Treg phenotype in patients with rheumatoid arthritis. Ann. Rheum. Dis. 74, 1265–1274 (2015).
Sonkoly, E. et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J. Allergy Clin. Immunol. 126, 581.e1-20–589.e1-20 (2010).
Ghaebi, M. et al. T cell subsets in peripheral blood of women with recurrent implantation failure. J. Reprod. Immunol. 131, 21–29 (2019).
Winger, E. E., Reed, J. L. & Ji, X. First-trimester maternal cell microRNA is a superior pregnancy marker to immunological testing for predicting adverse pregnancy outcome. J. Reprod. Immunol. 110, 22–35 (2015).
Pineles, B. L. et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am. J. Obstet. Gynecol. 196, 261.e1–261.e6 (2007).
Johansson, M., Bromfield, J. J., Jasper, M. J. & Robertson, S. A. Semen activates the female immune response during early pregnancy in mice. Immunology 112, 290–300 (2004).
Read, S., Malmstrom, V. & Powrie, F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J. Exp. Med. 192, 295–302 (2000).
Takahashi, T. et al. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J. Exp. Med. 192, 303–310 (2000).
Zhao, H. et al. Unique roles of infiltrating myeloid cells in the murine uterus during early to midpregnancy. J. Immunol. 194, 3713–3722 (2015).
Prins, J. R. et al. Unstable Foxp3+ regulatory T cells and altered dendritic cells are associated with lipopolysaccharide-induced fetal loss in pregnant interleukin 10-deficient mice. Biol. Reprod. 93, 95 (2015).
Bizargity, P., Del Rio, R., Phillippe, M., Teuscher, C. & Bonney, E. A. Resistance to lipopolysaccharide-induced preterm delivery mediated by regulatory T cell function in mice. Biol. Reprod. 80, 874–881 (2009).
Robertson, S. A., Care, A. S. & Skinner, R. J. Interleukin 10 regulates inflammatory cytokine synthesis to protect against lipopolysaccharide-induced abortion and fetal growth restriction in mice. Biol. Reprod. 76, 738–748 (2007).
Heimberg, A. M., Sempere, L. F., Moy, V. N., Donoghue, P. C. & Peterson, K. J. MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl Acad. Sci. USA 105, 2946–2950 (2008).
Vigorito, E., Kohlhaas, S., Lu, D. & Leyland, R. miR-155: an ancient regulator of the immune system. Immunol. Rev. 253, 146–157 (2013).
Zhang, C., Zhao, J. & Deng, H. 17beta-estradiol up-regulates miR-155 expression and reduces TP53INP1 expression in MCF-7 breast cancer cells. Mol. Cell. Biochem. 379, 201–211 (2013).
Zhao, J.-x, Zeng, Y.-y & Liu, Y. Fetal alloantigen is responsible for the expansion of the CD4+CD25+ regulatory T cell pool during pregnancy. J. Reprod. Immunol. 75, 71–81 (2007).
Sanchez-Diaz, R. et al. Thymus-derived regulatory T cell development is regulated by C-type lectin-mediated BIC/microRNA 155 expression. Mol. Cell. Biol. 37, e00341 (2017).
Yadav, M., Stephan, S. & Bluestone, J. A. Peripherally induced tregs - role in immune homeostasis and autoimmunity. Front. Immunol. 4, 232 (2013).
Tang, A. L. et al. CTLA4 expression is an indicator and regulator of steady-state CD4+ FoxP3+ T cell homeostasis. J. Immunol. 181, 1806–1813 (2008).
Wing, K., Yamaguchi, T. & Sakaguchi, S. Cell-autonomous and -non-autonomous roles of CTLA-4 in immune regulation. Trends Immunol. 32, 428–433 (2011).
O’Connell, R. M. et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 (2010).
Bayer, A. L., Yu, A. & Malek, T. R. Function of the IL-2R for thymic and peripheral CD4+CD25+ Foxp3+ T regulatory cells. J. Immunol. 178, 4062–4071 (2007).
Seddiki, N., Swaminathan, S., Phetsouphanh, C. & Kelleher, A. D. miR-155 is differentially expressed in Treg subsets, which may explain expression level differences of miR-155 in HIV-1 infected patients. Blood 119, 6396–6397 (2012).
Ceppi, M. et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl Acad. Sci. USA 106, 2735–2740 (2009).
Ohl, L. et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 21, 279–288 (2004).
Comerford, I. et al. A myriad of functions and complex regulation of the CCR7/CCL19/CCL21 chemokine axis in the adaptive immune system. Cytokine Growth Factor Rev. 24, 269–283 (2013).
Elmesmari, A. et al. MicroRNA-155 regulates monocyte chemokine and chemokine receptor expression in rheumatoid arthritis. Rheumatology (Oxford) 55, 2056–2065 (2016).
Moldenhauer, L. M., Keenihan, S. N., Hayball, J. D. & Robertson, S. A. GM-CSF is an essential regulator of T cell activation competence in uterine dendritic cells during early pregnancy in mice. J. Immunol. 185, 7085–7096 (2010).
Care, A. S., Ingman, W. V., Moldenhauer, L. M., Jasper, M. J. & Robertson, S. A. Ovarian steroid hormone-regulated uterine remodeling occurs independently of macrophages in mice. Biol. Reprod. 91, 60 (2014).
MartIn-Fontecha, A. et al. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J. Exp. Med. 198, 615–621 (2003).
Gunn, M. D. et al. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J. Exp. Med. 189, 451–460 (1999).
Hintzen, G. et al. Induction of tolerance to innocuous inhaled antigen relies on a CCR7-dependent dendritic cell-mediated antigen transport to the bronchial lymph node. J. Immunol. 177, 7346–7354 (2006).
Worbs, T. et al. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J. Exp. Med. 203, 519–527 (2006).
Dufour, J. H. et al. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 168, 3195–3204 (2002).
Harrison, E. B. et al. Induction of miR-155 after brain injury promotes type 1 interferon and has a neuroprotective effect. Front. Mol. Neurosci. 10, 228 (2017).
Dimitriadis, E., White, C. A., Jones, R. L. & Salamonsen, L. A. Cytokines, chemokines and growth factors in endometrium related to implantation. Hum. Reprod. Update 11, 613–630 (2005).
Wang, X. et al. MicroRNA-155-5p is a key regulator of allergic inflammation, modulating the epithelial barrier by targeting PKIalpha. Cell Death Dis. 10, 884 (2019).
Kong, W. et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell Biol. 28, 6773–6784 (2008).
Jasper, M. J. et al. Macrophage-derived LIF and IL1B regulate alpha(1,2)fucosyltransferase 2 (Fut2) expression in mouse uterine epithelial cells during early pregnancy. Biol. Reprod. 84, 179–188 (2011).
Sakaguchi, S., Vignali, D. A., Rudensky, A. Y., Niec, R. E. & Waldmann, H. The plasticity and stability of regulatory T cells. Nat. Rev. Immunol. 13, 461–467 (2013).
Tilburgs, T. et al. Fetal-maternal HLA-C mismatch is associated with decidual T cell activation and induction of functional T regulatory cells. J. Reprod. Immunol. 82, 148–157 (2009).
Wilson, R. L. et al. Vitamin D receptor gene ablation in the conceptus has limited effects on placental morphology, function and pregnancy outcome. PLoS ONE 10, e0131287 (2015).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Jimenez-Carretero, D., Ligos, J. M., Martinez-Lopez, M., Sancho, D. & Montoya, M. C. Flow cytometry data preparation guidelines for improved automated phenotypic analysis. J. Immunol. 200, 3319–3331 (2018).
Babicki, S. et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153 (2016).
Acknowledgements
This study was supported by Project Grant APP1041335 (to S.A.R. and S.C.B.) and Early Career Fellowship APP1092191 (to A.S.C.) from the National Health and Medical Research Council (Australia), and Discovery Grant DP160102366 (to S.A.R.) from the Australian Research Council.
Author information
Authors and Affiliations
Contributions
S.A.R. and J.E.S. designed and supervised the study. L.M.M., J.E.S., B.Z., A.S.C. and H-Y.C. performed the experiments. J.E.S., L.M.M., A.S.C. and C.M.H. analyzed the data. S.C.B. contributed to the project design and discussions on data interpretation. S.A.R and J.E.S. wrote the manuscript. All authors revised drafts and reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Schjenken, J.E., Moldenhauer, L.M., Zhang, B. et al. MicroRNA miR-155 is required for expansion of regulatory T cells to mediate robust pregnancy tolerance in mice. Mucosal Immunol 13, 609–625 (2020). https://doi.org/10.1038/s41385-020-0255-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-020-0255-0
This article is cited by
-
Investigation into the role of the MITA-TRIM38 interaction in regulating pyroptosis and maintaining immune tolerance at the maternal-fetal interface
Cell Death & Disease (2023)
-
Recent highlights in the immunomodulatory aspects of Treg cell-derived extracellular vesicles: special emphasis on autoimmune diseases and transplantation
Cell & Bioscience (2022)
-
Transcriptomic analysis of the seminal vesicle response to the reproductive toxicant acrylamide
BMC Genomics (2021)
-
Adversity in early life and pregnancy are immunologically distinct from total life adversity: macrophage-associated phenotypes in women exposed to interpersonal violence
Translational Psychiatry (2021)
-
Toll-like receptor-4 null mutation causes fetal loss and fetal growth restriction associated with impaired maternal immune tolerance in mice
Scientific Reports (2021)