Abstract
Epithelial cells of the conducting airways are a pivotal first line of defense against airborne pathogens and allergens that orchestrate inflammatory responses and mucociliary clearance. Nonetheless, the molecular mechanisms responsible for epithelial hyperreactivity associated with allergic asthma are not completely understood. Transcriptomic analysis of human airway epithelial cells (HAECs), differentiated in-vitro at air-liquid interface (ALI), showed 725 differentially expressed immediate-early transcripts, including putative long noncoding RNAs (lncRNAs). A novel lncRNA on the antisense strand of ICAM-1 or LASI was identified, which was induced in LPS-primed HAECs along with mucin MUC5AC and its transcriptional regulator SPDEF. LPS-primed expression of LASI, MUC5AC, and SPDEF transcripts were higher in ex-vivo cultured asthmatic HAECs that were further augmented by LPS treatment. Airway sections from asthmatics with increased mucus load showed higher LASI expression in MUC5AC+ goblet cells following multi-fluorescent in-situ hybridization and immunostaining. LPS- or IL-13-induced LASI transcripts were mostly enriched in the nuclear/perinuclear region and were associated with increased ICAM-1, IL-6, and CXCL-8 expression. Blocking LASI expression reduced the LPS or IL-13-induced epithelial inflammatory factors and MUC5AC expression, suggesting that the novel lncRNA LASI could play a key role in LPS-primed trained airway epithelial responses that are dysregulated in allergic asthma.
Similar content being viewed by others
Introduction
Allergic asthma is a heterogenous chronic respiratory disease characterized by airway mucosal remodeling and hyperreactivity, and during an exacerbation is symptomatically manifested as episodes of wheezing, productive cough, and dyspnea.1 Genetic predisposition and the environmental factors, such as exposure to airborne allergens, pathogenic microbes or their byproducts, and strenuous exercise, can trigger a dysregulated TH2 immune response in susceptible asthmatics.2,3 Current treatment regimens targeting the well-characterized cellular and molecular immunomodulators including inhaled corticosteroids, β2-agonists, anti-IgE antibodies, mast cell stabilizers, and leukotriene modifiers have been effective in managing the mild-to-moderate allergic asthma but has demonstrated limited efficacy in controlling the severe asthma, and, if not managed, could perpetuate resistance.2 Therefore, greater research efforts are needed to improve upon the current understanding on the molecular basis of allergic asthma and pathophysiology of airway hyperreactivity.
Airway epithelial cells (AECs) of the respiratory tract play a significant role in allergic asthma.4,5,6,7 AECs help mitigate the inhaled airborne allergens/pathogens via mucociliary clearance mechanisms and orchestrate the airway mucosal immune responses.5 However, chronic lung injury, repair, and regeneration leads to morphological and functional anomalies that result in a mucus hypersecretory state, leading to defective mucociliary clearance and mucus plugging of small airways or broncho-obstruction.8 The resulting pathological condition is also referred to as chronic mucus hypersecretion and is associated with the asthma severity.6,7,8,9 The polymeric gel-forming mucins, MUC5AC and MUC5B, are the most abundant secretory mucins of AECs that are genetically regulated by the transcriptional factors, SPDEF and STAT6, which are critical players in the regulation of TH2 immune responses of inflammatory cytokines, like IL-4, IL-5, and IL-13.6,7,8
Repetitive environmental exposures to allergens, pathogens, or their byproducts, evoke an immunological memory to establish host immunity. Besides lymphocytes, innate immune cells, including monocytes, macrophages, and natural killer (NK) cells, have also been shown to preserve an immunological memory and respond in a host-beneficial manner, which has been termed as a ‘trained immunity’.10 The mechanisms involved in this trained immunity include epigenetic and transcriptomic modifications as well as metabolic and functional reprogramming.11 AECs are crucial innate immune cells that orchestrate lung immunity.12,13,14 Nonetheless, the studies establishing trained immunity in these cells are limited.15 Recently, we analyzed animal models of epithelial trained immunity, whereby, rodents previously primed with LPS responded with a rapid mucous cell hyperplasia to a secondary LPS challenge that involved spatiotemporal upregulation of TLR4 and EGFR.16 Therefore, we investigated whether human AECs (HAECs) also respond in a memory-dependent manner, and probed for the underlying genetic or transcriptomic programming mechanism(s) associated with the allergic asthma.
More than two-thirds of human transcriptome consists of noncoding RNA species, including long noncoding RNAs (lncRNAs). LncRNAs are transcripts comprised of over 200 nucleotides that lack protein coding potential and can be found in the nuclear or cytosolic regions. According to their biogenesis, lncRNAs are categorized into intergenic, intronic, natural antisense transcripts, bidirectional, or enhancer lncRNAs.17 LncRNAs are mainly involved in essential cellular functions like chromatin modification, mRNA decay, alternative splicing, and regulation of transcription by cis (in the nucleus) or trans (in the cytoplasm) mechanisms. LncRNAs exhibit tissue-specific expression patterns that dictate their specific function in a particular type of cell or tissue.18,19 Recently, several studies reported on lncRNAs associated with lung diseases including allergic asthma, but only a few of them had been experimentally validated.20,21 In the present study, we performed a high-throughput transcriptomic analysis to identify novel lncRNAs that might be one of the immediate-early immunomodulators of HAECs. The role of this novel lncRNA was investigated in airway tissues, and HAECs of asthmatic and control subjects using qPCR, multispectral immunostaining, fluorescent in-situ hybridization (FISH), and by cell fractionation analyses; and the RNA interference-mediated gene silencing was employed to probe the lncRNA’s role in HAEC inflammatory and mucous responses.
Results
LPS-primed human AECs preserve a memory-dependent rapid mucous response
To determine whether primary HAECs respond in a memory-dependent manner to LPS exposure, HAECs were differentiated for 3 weeks at the air-liquid interface (ALI) on transwells. Separate transwells of well-differentiated HAECs were then either primed with 100 ng/ml LPS (L) or left unprimed (U). On day 10, half of the transwells from each group (L or U) were challenged with 0 or 10 ng/ml LPS; resulting in four treatment groups identified as L/0 and L/10 or U/0 and U/10, respectively. The LPS-priming augmented a robust mucous phenotype at 24 h post-secondary LPS challenge (L/10), as assessed by the expression levels of secretory mucin MUC5AC (Fig. 1a), and SPDEF (Fig. 1b), a master regulator of the airway mucous response.22 Among LPS-primed cells, MUC5AC mRNA levels were more than twofold higher in L/0 group and were induced >10-fold following LPS challenge (L/10 group) compared to controls (U/0 or U/10 group). SPDEF mRNA levels were also more than fourfold induced by LPS challenge in L/10 cells. There were no discernable changes in MUC5AC or SPDEF expression by 10 ng/ml LPS treatment in unprimed controls (U/10).
ALI-differentiated HAECs on transwells were either treated with 100 ng/ml LPS (L) or were left untreated (U), and on d 10 post first treatment, half of the transwells from each group were challenged with 0 or 10 ng/ml LPS, resulting in four treatment groups identified as L/0 and L/10 or U/0 and U/10, respectively. Relative quantities of MUC5AC (a) and SPDEF (b) mRNA levels in U/0, L/0, U/10, and L/10 HAECs as determined by qRT-PCR at 24 h post last treatment. c Micrographs of differentiated HAECs stained for β-tubulin (green, ciliated cells) and MUC5AC (red); and DAPI-stained nuclei (blue) from U/0, L/0, U/10, and L/10 HAECs, scale - 5 μ. d Quantification of MUC5AC + HAECs in each treatment group. Data shown as mean ± SEM of three independent experiments (n = 6/gp); *p < 0.05; **p < 0.01 as analyzed by the one-way ANOVA and Tukey’s multiple comparison test.
MUC5AC protein expression was analyzed by immunostaining and cells were co-stained for β-tubulin expression to identify the ciliated cells (Fig. 1c). The percentage of MUC5AC immunopositive cells was significantly higher (>3-fold) in the LPS-primed L/10 group compared to unprimed controls (Fig. 1d). There was no apparent effect on the ciliated cell population with 38.8 ± 2.8% in L/10 and 40.1 ± 4.1% in the U/10 group. Thus, the in-vitro differentiated and LPS-primed HAECs demonstrated a memory-based rapid mucous response.
A long noncoding RNA species implicated in LPS-primed memory response
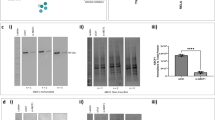
To determine the molecular mechanism involved in this memory-based hyperreactive or ‘trained’ mucous response, well-differentiated primary HAECs were treated with LPS (100 ng/ml) for 2 h to analyze the immediate-early responses. Total RNA was isolated, and transcriptomic analysis was performed following high-throughput RNA-sequencing as described in the methods section. The principal component analysis of the RNA seq data showed distinct clustering of the controls and the experimental LPS-treated samples (Fig. 2a). Further in-silico analysis of the RNA-seq data helped identify 725 differentially expressed transcripts that were statistically significant (Fig. 2b). As expected, the transcriptomic changes were similar to that of a classical proinflammatory response with significant changes in gene expression related to the immune function, cell proliferation, and remodeling (Supplementary Fig. S1A and Table S1). Pathway, Gene Ontology: Biological Process, and PubMed literature co-citation analysis identified 1404 biological processes, and 104 different pathways that were significantly affected (Supplementary Fig. S1B, C; Tables S2 and S3).
Primary HAECs were differentiated at the air-liquid interface and were treated with LPS (100 ng/ml) for 2 h. Total RNA was isolated from cells and RNA-Seq analysis was performed. a PCA analysis of the RNA seq data showing the distinct grouping of control and LPS-treated (experimental) samples. b Volcano plot analysis of RNA seq data showing the distribution of transcripts that were upregulated or downregulated following LPS treatment. c Genetic map of lncRNA LASI encoded on the antisense strand of ICAM1 within chromosome 19. d Relative quantities of LASI transcript levels in U/0, L/0, U/10, and L/10 HAECs as determined by qRT-PCR. Data shown as mean ± SEM with n = 3–6/group; *p < 0.05; **p < 0.01 as analyzed by the one-way ANOVA and Tukey’s multiple comparison test.
We focused on the long noncoding RNAs or lncRNAs because they have been shown to modulate gene expression and cellular functions at transcriptomic and epigenetic levels by regulating chromatin modification and mRNA stability.18,19 Several putative lncRNA transcripts were expressed differentially following LPS challenge (Table 1), and we selected lncRNA AC011511.2, a lncRNA on the antisense strand of ICAM-1 or LASI for further analysis because ICAM-1 plays a critical role in airway inflammation and remodeling.23 In addition, ICAM-1 expression itself is associated with LPS responses, airway inflammation, and asthma.24 Sequencing and annotation analysis revealed that this LASI transcript is a 32.4 Kb long with 3 exons and 2 introns (Fig. 2c and see Supplementary Fig. S2 for a detailed map). This lncRNA showed a negligible protein coding potential with a Fickett Testcode score of 0.27327 and a coding probability of 0.197913 (http://cpc2.cbi.pku.edu.cn).
Next, to validate the RNA-Seq data, we analyzed the expression of LASI transcripts in the in-vitro model of trained mucous response (Fig. 1). Among LPS-primed cells, LASI transcript levels were more than twofold higher in L/0 group and the secondary LPS challenge further augmented the LASI transcript levels by more than sixfold in L/10 group compared to unprimed controls (Fig. 2d), implicating the lncRNA LASI in LPS-primed mucous response.
Airway mucous cell hyperplasia and elevated LASI transcript levels in allergic asthmatics
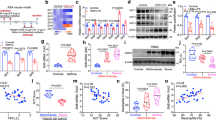
Next, we investigated the LASI transcripts in the airway tissues of patients with a reported history of asthma. Archived airway sections from self-reported asthmatics and control subjects were analyzed for airway epithelial mucin load by Alcian blue (AB) staining with H&E contrast (Fig. 3a). The demographics of the subjects analyzed is shown in Table 2. Based on the histological quantitation, there were more than fivefold higher AB + mucous cells (MCs) per mm basal lamina (BL) in asthmatics’ airways compared to non-asthmatic controls (Fig. 3b), and there was increased eosinophilic infiltration in the airway epithelium of asthmatics (Fig. 3c). MUC5AC immunostaining (Fig. 3d) and the imaging analysis showed >10-fold higher MUC5AC immunopositivity in asthmatic airways per mm of BL (Fig. 3e). Among transcript levels, there was significantly elevated (>10-fold) MUC5AC (Fig. 3f) and SPDEF (Fig. 3g) mRNA levels in asthmatics that also had more than fivefold higher expression of LASI transcripts (Fig. 3h).
a Representative micrographs of airway sections of control and asthmatic subjects stained with Alcian blue for mucopolysaccharides and contrasted with H&E staining, scale – 5 μ. Quantification of AB + mucous cells (b) and eosinophils (c) per mm of basal lamina (BL). d Micrographs of airway sections stained for MUC5AC (green) and the DAPI-stained nuclei (blue) from control and asthmatic subjects, scale – 5 μ. e Quantification of MUC5AC + mucous cells per mm of BL in airways of controls and asthmatics. Relative quantities of MUC5AC (f), SPDEF (g), and LASI (h) in asthmatic HAECs compared to controls. i Micrographs of FISH-stained airway sections from control and asthmatic subjects showing LASI transcripts (red) and co-stained for MUC5AC (green) along with the DAPI-stained nuclei (blue), scale – 5 μ. Lower panels show the magnified image of the epithelial cells showing LASI transcripts in the inset drawn on the upper panels. j Quantification of LASI transcript per epithelial cell in airways of controls and asthmatics. Data shown as mean ± SEM; n = 6–7/gp; *p < 0.05; **p < 0.01; ****p < 0.0001 as analyzed by student’s t test.
To analyze the airway epithelial-specific LASI expression, we performed RNA FISH (RNAScope®, Advanced Cell Diagnostics, USA) and sections were co-immunostained for MUC5AC mucin. LASI expression was observed in MUC5AC + goblet cells as well as in other epithelial cells (Fig. 3i). The weighted distribution (H-score) of LASI expression was twofold or higher in airway epithelium of asthmatics compared to non-asthmatic controls (Fig. 3j). Together, these data suggest that LASI expression in airway epithelia is strongly associated with mucous cell hyperplasia in asthmatics.
Ex-vivo grown asthmatic HAECs show hyperreactive mucin and LASI expression
Primary HAECs from asthmatic and non-asthmatic control subjects were differentiated on transwells and analyzed for the LPS-primed memory response as described in Fig. 1. At 24 h post LPS treatment, there was significantly increased expression of MUC5AC (Fig. 4a) and SPDEF (Fig. 4b) mRNAs in LPS-primed HAECs from asthmatic subjects compared to that of controls and expression was higher even at the baseline (U/0) among asthmatic HAECs. The LASI transcripts also showed elevated expression among asthmatic HAECs, and LPS priming further augmented the expression levels (Fig. 4c). These data corroborate that LASI expression is strongly associated with the LPS-primed hyperreactive mucin expression of asthmatic HAECs.
Transwells of differentiated HAECs from controls and asthmatics were either primed with 100 ng/ml LPS (L) or were left untreated (U), and on d 10, half of the transwells from each group were challenged with 0 or 10 ng/ml LPS, resulting in four treatment groups identified as L/0 and L/10 or U/0 and U/10, respectively. Relative quantities of MUC5AC (a), SPDEF (b), and LASI (c) transcript levels in U/0, L/0, U/10, and L/10 HAECs from controls and asthmatics as determined by qRT-PCR at 24 h post last treatment. Data shown as mean ± SEM; n = 4/gp; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, compared to U/0 controls or as indicated based on the two-way ANOVA and Tukey’s multiple comparison test.
LPS and IL-13 induce nuclear-enriched LASI expression, but IL-13 primed cells do not sustain a memory-dependent response
Next, we followed the expression of LASI transcripts in normal HAECs that were treated with 100 ng/ml LPS for 0, 0.5, 2, 6, 12, and 24 h. The LASI transcript levels were induced as early as 0.5 h post LPS treatment with the highest expression observed at 2 and 6 h post-treatment, and the expression levels remain elevated even at 24 h post-treatment (Fig. 5a). The mRNA levels of the mucin regulatory transcription factor SPDEF showed increased expression starting at 2 h post-treatment with the highest expression at 24 h post-treatment (Fig. 5b).
Quantification of LASI (a) and SPDEF (b) transcript levels in HAECs at 0, 0.5, 2, 6, 12, and 24 h following the treatment with 100 ng/ml LPS. Quantification of LASI (c) and SPDEF (d) transcript levels in HAECs at 0, 0.5, 2, 6, 12, and 24 h following the treatment with 10 ng/ml IL-13. Relative quantities of LASI (e) and SPDEF (f) transcripts in nuclear and cytosolic fractions of LPS and IL-13 treated HAECs compared to non-treated (NT) cells. Data shown as mean ± SEM; n = 4–6/gp; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, based on the one-way ANOVA and Dunnett’s multiple comparison test for (a–d), and for (e and f), compared to NT cytosolic levels based on one-way ANOVA and Tukey’s multiple comparison test.
Using airway organ cultures, we have recently shown that IL-13 is an important inflammatory factor that mediates mucin expression in response to the LPS challenge.25 IL-13 is a pleiotropic TH2 cytokine, which plays a central role in asthma pathogenesis and mucin expression.25,26 Therefore, we analyzed the effect of IL-13 on the LASI transcript levels and HAEC responses. HAECs were treated with 10 ng/ml IL-13 for 0, 0.5, 2, 6, 12 and 24 h, and there was significant induction in LASI transcript levels at 2 and 6 h post-IL-13 treatment with more than a sixfold increase at 2 h post-treatment (Fig. 5c). But the elevated transcript levels were not sustained beyond 6 h post-treatment, unlike LPS response (Fig. 5a). The SPDEF mRNA levels showed the highest expression at 2 h post-IL-13 treatment (Fig. 5d), and the expression levels were similar to that of uninduced cells (0 h) by 12 h post-treatment.
LncRNA functions are predominantly dictated by their subcellular localization, therefore, we investigated the subcellular distribution of LASI transcripts in HAECs. For this purpose, we isolated nuclear and cytosolic fractions from LPS, and IL-13 treated, and non-treated (NT) HAECs. In the uninduced NT cells, the LASI transcripts were enriched (>4-fold) in the nuclear fraction, and both nuclear and cytosolic fractions of LPS and IL-13 treated cells had induced LASI transcript levels with the nuclear fractions showing more than a 25-fold increase (Fig. 5e). A similar change in the SPDEF mRNA levels was observed, however, both nuclear and cytosolic fractions had a comparable induction (>5-fold) in the treated cells (Fig. 5f). Thus, these data suggest that the LPS and IL-13 induced LASI transcripts are predominantly localized and enriched in the nuclear region of HAECs.
We next examined whether IL-13 primed cells also preserve a memory-based augmented response. The differentiated HAECs were primed with 10 ng/ml IL-13 or left unprimed (U) and on d 10 were treated with IL-13, with some groups primed with 100 ng/ml LPS as positive controls. IL-13 treatment induced the LASI transcripts but there was no difference in unprimed (U/IL-13) and the IL-13-primed (IL-13/IL-13) or LPS-primed (L/IL-13) cells (supplementary Fig. S3A). A similar trend was observed in SPDEF mRNA levels with no difference in unprimed (U/IL-13) and the IL-13-primed (IL-13/IL-13) HAECs (supplementary Fig. S3B). However, following IL-13 treatment, LPS primed cells showed significant induction in SPDEF mRNA levels compared to unprimed or IL-13-primed cells.
Nuclear-enriched LASI transcripts associated with increased epithelial inflammatory factors
In order to further assess the association of LASI transcripts with inflammatory and mucous responses, differentiated HAECs were also analyzed for LASI transcripts localization using RNAscope® FISH analysis. LASI transcripts were mostly enriched in the perinuclear region at 2 h post-LPS treatment (Fig. 6a) with twofold or higher LASI transcript expression following LPS treatment compared to non-treated (NT) controls (Fig. 6b). LPS treatment resulted in increased expression of ICAM-1 (Fig. 6c), CXCL-8 (Fig. 6d), and IL-6 (Fig. 6e) mRNAs compared to controls.
a Micrographs of HAECs showing RNA FISH stained individual LASI transcripts (red) using RNAscope® technology and DAPI-stained nuclei (blue) in LPS-treated and non-treated (NT) HAECs, scale – 5 μ. b Quantification of the number of LASI transcripts per cell (denoted as H-score, see Methods for details) in LPS-treated and non-treated HAECs. Relative quantities of ICAM-1 (c), CXCL-8 (d), and IL-6 (e) mRNA levels in LPS-treated HAECs as determined by qRT-PCR at 24 h post-treatment in comparison with the non-treated controls. Data shown as mean ± SEM; n = 3–7/gp; *p < 0.05; ***p < 0.001 as analyzed by student’s t test.
A similar trend was observed in the cells treated with IL-13 (supplementary Fig. S4), with an increased aggregation of LASI transcripts in the perinuclear region (Fig. S4A) and >2-fold increase in LASI transcripts in IL-13-treated cells than controls (Fig. S4B). In addition, LPS treatment induced the ICAM-1 (Fig. S4C) and CXCL-8 (Fig. S4D), however, there was no change in IL-6 (Fig. S4E) expression. Thus, the increased LASI transcripts and observed enrichment in the nuclear/perinuclear region may be one of the initial responses in the onset of airway mucous and inflammatory responses.
Blocking LASI expression suppresses LPS and IL-13 induced HAEC responses
Next, we interrogated the role of LASI by blocking its expression using siRNA interference technology. Primary HAECs were transfected with siRNA targeting LASI transcripts (siLASI) or with non-specific siRNA control (siCTRL) and challenged with LPS (100 ng/ml). LASI transcript levels were completely blocked in the HAECs pretreated with siLASI compared to siCTRL cells with no discernible effect of LPS treatment (Fig. 7a). LPS-induced MUC5AC mRNA expression was attenuated in siLASI-transfected cells compared to siCTRL cells (Fig. 7b). Among epithelial cell secreted factors, the siLASI-treated cells showed attenuation of LPS-induced expression of ICAM-1 (Fig. 7c) and IL-6 (Fig. 7d) mRNAs. However, there was no change in CXCL-8 mRNA levels (Fig. 7e), indicating an indirect role of LASI in LPS-mediated CXCL-8 induction. In another set of experiments, siLASI-treated HAECs showed attenuation of the IL-13 induced changes in LASI, MUC5AC, SPDEF, ICAM-1, and CXCL-8 transcript expression compared to siCTRL cells (supplementary Fig. S5). Taken together, these data suggest that LASI transcripts may be an important regulator of inflammatory and mucous responses of HAECs.
Primary HAECs grown in submerged culture conditions were transfected with siRNA targeting LASI transcripts (siLASI) or with control non-specific siRNAs (siCTRL) and at 24 h post-transfection cells were challenged with 100 ng/ml LPS and cells were analyzed at 24 h post-treatment. Relative quantities of LASI transcripts (a), and MUC5AC (b), ICAM-1 (c), IL-6 (d), and CXCL-8 (e) mRNA levels in LPS-treated HAECs transfected with siLASI-compared to the siCTRL-transfected cells. Data shown as mean ± SEM; n = 3/gp; *p < 0.05; **p < 0.01; ***p < 0.001, compared to siCTRL group or as indicated based on one-way ANOVA and Tukey’s multiple comparison test.
Discussion
In line with our recently reported in-vivo observation in animal models of trained airway immunity,16 the present study corroborates that LPS-primed differentiated HAECs preserve a memory-based airway hyperreactivity that is associated with allergic asthma and identified the role of a novel lncRNA LASI therein. In HAECs, LPS or IL-13 mediated induction of LASI transcript levels elevate rapidly in the nuclear/perinuclear region and possibly regulate SPDEF levels to augment HAEC inflammatory and mucous responses. Most importantly, blocking the expression of LASI by small interfering RNA-mediated silencing attenuated inflammatory and mucous responses, indicating of its importance in airway epithelial biology.
A number of cutting-edge studies have now widely established the innate memory-based responses or ‘trained immunity’ of innate immune cells, which primarily involves epigenetic reprogramming via DNA methylation, microRNAs, and/or long noncoding RNAs.12,13 For example, studies investigating the European and U.S. farm and non-farm children’s pulmonary health show that early life exposure to airborne immunomodulators protects individuals against allergic asthma by activating a form of trained immunity in the lung mucosa and/or innate immune cells.27,28,29 Although, this trained immunity primarily shows beneficial effects; pan-protection and enhanced immune responses could also lead to the development of chronic disorders,11 as reported here too. Nonetheless, this immunological memory-dependent phenomenon offers a novel way to identify potential intervention targets that can readily modulate the downstream mediators and offer a novel benchmark for the personalized medicine approach in managing allergic asthma.
In the present study, we showed that LPS-primed differentiated HAECs preserve a memory-dependent rapid recall response following the LPS challenge. Transcriptomic analysis showed that a novel lncRNA LASI could be responsible for this trained response as an immediate-early transcript primed by LPS signaling pathways. The expression pattern of LASI transcripts in the in-vitro model of trained airway response supported their significant role in aggravated inflammation and mucous cell hyperplasia. Both qPCR and RNA-FISH experimental analysis showed a robust LASI expression in allergic asthmatic airway tissues and depicted a strong association with mucin hyperexpression, as confirmed ex-vivo in asthmatic HAECs. Interestingly, the basal expression levels of LASI, MUC5AC, and SPDEF were higher than the control HAECs from healthy subjects, and upon secondary challenge, these levels increased exponentially. Although many HAEC-associated lncRNAs have been identified recently,30,31 however, there are limited studies implicating lncRNAs of airway epithelial origin. Most of the reported asthma-associated lncRNAs have been either identified in peripheral blood mononuclear cells,20,32 or in the airway smooth muscle cells.33,34 In contrast, our present study is the first report on the conducting airway epithelial-specific lncRNA that is associated with allergic asthma and responds in a memory-dependent manner with a robust immediate-early expression and nuclear enrichment.
The impact of lncRNAs on cell functions are primarily governed by the subcellular localization, whereby, they can modulate chromatin modifications, mRNA stability, alternative mRNA processing, and transcription.18,19 Most of the lncRNAs accumulate in the nucleus and act via cis- or trans-mechanisms for regulating gene expression, while a very few lncRNAs translocate into the cytosol and act via trans-mechanisms.35 During pathological conditions, lncRNAs expression and localization could be altered, affecting their role in pre- and post-transcription processes by modulating interactions with genomic DNA, mRNA, miRNA, and various regulatory proteins and lipids.17,36 In the present study, analysis of the subcellular fractions revealed that LASI transcript levels were induced and accumulated in nuclear/perinuclear regions. Interestingly, a mucin regulatory transcription factor SPDEF was also induced in the LPS-primed cells and mirrored the expression patterns of LASI in all experimental conditions, including memory-dependent trained mucous response and the expression kinetics. SPDEF regulates the growth and differentiation of mucus-producing goblet cells as well as expression of secretory MUC5AC mucin and is expressed in very high levels in asthmatic patients than healthy controls.5,8,22 In cells with blocked LASI expression, the LPS or IL-13 induced MUC5AC and SPDEF expression were attenuated indicating the role for LASI in mediating the molecular regulation of mucous response. Detailed studies are currently underway investigating the mechanisms for LASI-mediated upregulation of MUC5AC, and SPDEF during hyperreactive responses.
In contrast to LPS priming, IL-13 primed HAECs are unable to preserve the observed immunological memory. Further investigation is needed to distinguish how the LPS-primed and TLR4 and downstream signal transducers are able to sustain the LASI expression and associated recall responses while the IL-13 primed, and STAT6-mediated transducers are not. This could also be an innate mechanism by which lncRNA-related regulation differ the response to a cytokine vs pathogen. In addition, the findings of LPS-mediated induction of ICAM-1 and LASI also unravel a unique correlation of expression between genes encoded on the opposite strands and also demands further molecular characterization of the epigenetic modifications and bidirectional transcription mechanisms in the current experimental settings, which are beyond the scope of the current study.
HAECs synthesize and secrete various inflammatory factors to help orchestrate the lung mucosal immune response by recruiting and activating various immune cells.37 Among AEC-derived inflammatory factors, IL-6 and CXCL-8 have been predominantly observed in chronic lung diseases including allergic asthma. These are pleiotropic immunomodulators, which regulate adhesion molecules like ICAM-1 and act as chemoattractants to recruit effector immune cells like monocytes and lymphocytes.38,39 Therefore, controlling the expression or function of these inflammatory mediators could also help in reducing the disease severity. Recent seminal studies revealed that lncRNAs, IL17AS and MIR3142HG regulate IL-6 and CXCL-8 expression via NF-κB pathways in IPF tissues;40 lncRNA PVT1 regulates IL-6 and CXCL-8 release in ASMCs;21,41 and lncRNA ANRIL regulates IL-6, IL-13, IL-4, and IL-17 in allergic rhinitis condition.42 In the present study, HAECs treated with LPS/IL-13 showed increased IL-6, CXCL-8, and ICAM-1 expression, and blocking LASI expression via siRNA attenuated IL-6 and ICAM-1 mRNA expression but not CXCL-8. These studies thus indicate that LASI transcripts regulate the expression of HAEC inflammatory mediators.
There are a few limitations in the current study that should be considered while drawing conclusions. Firstly, the number of subjects analyzed in this study is very low due to the non-availability of well-preserved airway tissues from allergic asthma and non-asthmatic subjects. This is also reflected in the primary HAECs analyzed from healthy or asthmatic donors. In addition, three of the asthmatics had a smoking history which could be a confounding factor in the observed effects. However, we have analyzed airway tissues from smokers with no chronic lung disease history, and LASI and mucin expression levels were similar to those observed in controls (personal observation). Also, due to the limitations of the amount of samples/transwells for differentiated HAECs, the subcellular fractionation was carried out using cells from submerged culture conditions.
Conceptually, the form of ‘pathogenic trained response’ of asthmatic HAECs observed here is in total contrast to the development of ‘host-beneficial trained response’ to LPS/endotoxins or farm dust as reported in the developing lungs, where the early life exposure to immunomodulators protects against allergic asthma.27,28,29 This disparity could be due to the use of adult HAECs in this study, and longitudinal analysis of the pediatric HAECs is needed to understand this dichotomy. In addition, the reported hyperreactive response of HAECs observed here should not be compared with the low dose allergen or LPS models that involve lymphocytes and other immune cells that augment a tolerogenic response and are the basis for the allergen immunotherapies. Currently, studies are underway to help identify and characterize the binding partners for LASI lncRNA. These ongoing and other future studies could help unravel the molecular regulatory mechanisms by which LASI lncRNA modulates airway epithelial responses and asthma pathophysiologies. In conclusion, this is a pioneering study reporting that the human lung AECs preserve a memory-based ‘trained’ immune response by upregulating lncRNA LASI expression and allergic asthma-associated airway hyperreactivity.
Methods
Human airway epithelial tissues and cells
Airway tissue specimens and primary human airway epithelial cells (HAECs) from subjects with and without asthma were obtained from deidentified organ donors through the Life Alliance Organ Recovery Agency of the University of Miami, according to protocols approved by the local Institutional Review Board. Tissues specimens were fixed in 10% neutral buffered formalin and conventionally processed for paraffin embedding and sectioning. Airway epithelial cells were isolated, and cultured on air-liquid interface (ALI) as previously described.43,44 Normal human primary AECs were also kindly provided by Dr. Scott Randell at the Marsico Lung Institute/Cystic Fibrosis Research Center at the University of North Carolina, Chapel Hill, USA. Lung tissues were procured under protocol #03–1396 approved by the University of North Carolina at Chapel Hill Biomedical Institutional Review Board.
Cell culture treatments
HAECs were maintained in bronchial epithelial growth medium (BEGM, Lonza, Walkersville, MD). For ALI culture, cells were plated onto collagen IV–coated 24 mm Transwell-clear culture inserts (Corning Costar Corporation, Cambridge, MA) at a density of 5 × 105 cells/cm2 in ALI media. The apical surface of the cells was exposed to air and cells were cultured for another 21 days until they were fully differentiated.43,44 HAECs were treated with LPS (Pseudomonas aeruginosa serotype 10, Sigma-Aldrich, St. Louis, MO) or with recombinant human IL-13 (Peprotech Ltd.). Following treatments, the membrane quarters were used for RNA and protein isolation or were fixed in 4% paraformaldehyde for immunostaining. For memory-based responses, differentiated HAECs were either primed with 100 ng/ml LPS (L) or left unprimed (U) and on day 10 post priming, half of the transwells from each group (L or U) were challenged with 0 or 10 ng/ml LPS; resulting in four treatment groups, namely, L/0 and L/10 or U/0 and U/10, respectively. All the methods were performed in accordance with the institutional guidelines and regulations at FIU.
Transcriptomic analysis
Illumina TruSeq Stranded Total RNAseq libraries with ribosomal depletion were prepared per manufacturer’s instructions (Illumina, Inc, San Diego, CA). Libraries were sequenced on a HiSeq2500 (2 × 100 PE reads; 50 million reads/sample) at HudsonAlpha Institute for Biotechnology, Huntsville, AL. RNA-Seq data was analyzed as described previously.45,46 Briefly, reads were aligned to the GRCh38.P10 reference genome with Hisat2 and assembled into transcripts with Cufflinks2. CuffDiff was utilized to determine differential expression. Raw sequencing data have been deposited into the Sequence Reads Archive, and the experimental data files placed in Gene Expression Omnibus (GEO). Long noncoding genes were identified by their accession numbers. The RNA-seq data have been deposited in NCBI’s GEO47 and are accessible at GEO accession number GSE139370.
Pathway analysis
ToppFun (https://toppgene.cchmc.org/) was utilized for pathway analysis as previously described.48 Briefly, significantly different gene names (HUGO Gene Nomenclature Committee names) were entered into the ToppGene Suite. Duplicate and unknown genes were removed. Gene Ontology biological process and pathway analysis were utilized to determine significantly different functional genomic changes. Fast Ward Hierarchical Clustering was performed using the Log2x + 1 transformation of the data in the JMP genomics 8.0 (SAS Institute, Cary, NC). A p value (<0.05 with FDR correction) of each annotation of a test gene is derived by random sampling from the whole genome.
Histochemical staining
Airway tissue sections (5 µm) were deparaffinized in xylene and hydrated in graded ethanol series and deionized water. Histochemical staining with either hematoxylin and eosin (H&E) or AB and Hematoxylin-Eosin reagent (AB-H&E) was carried out as described.49 Mucous/Goblet cells and the eosinophils per mm BL were determined by counting AB+ and Eosin+ polymorphonuclear cells, respectively.
Immunostaining and fluorescent imaging analysis
For immunohistochemical staining, deparaffinized and hydrated airway tissue sections and treated HAECs were washed in 0.05% v Brij-35 in PBS (pH 7.4) and immunostained as described previously.50 Briefly, antigens were unmasked by steaming sections in 10 mM Citrate buffer (pH 6.0) followed by incubation in a permeabilizing blocking solution, and sections were stained with antibodies to MUC5AC (Millipore Inc., MA), and β-tubulin (Cell Signaling Tech, MA) or isotype controls. The immunolabelled cells were detected using respective secondary antibodies conjugated flours (Jackson ImmunoResearch Lab Inc., West Grove, PA) and mounted with 4′,6-diamidino-2-phenylindole (DAPI) containing Fluormount-GTM (SouthernBiotech, Birmingham, AL) for nuclear staining. Immunofluorescent images were captured using BZX700 Microscopy system (Keyence) and analyzed using NIH Image J software.
Quantitative RT-PCR
Total RNA was isolated using RNAeasy kit (Qiagen, MD) as per manufacturer’s instructions, quantified using Synergy HTX reader (BioTek, VT) and cDNA were synthesized using iScript kit (BioRad, CA). MUC5AC, SPDEF, and LASI transcripts were quantified using FAM-based primer/probe sets (Applied Biosystems, ThermoFisher) using the TaqMan Gene expression kit (ThermoFisher), whereas, ICAM-1, IL-6, and CXCL-8 mRNA were quantified using SYBR green-based primers or iTaq Master Mix (BioRad, CA) in the Agilent Stratagene Mx3000P Real-Time PCR System (ThermoFisher). Relative quantities were calculated by normalizing averaged CT values to CDKN1B or β-Actin to obtain ΔCT, and the fold-change (ΔΔCT) over the controls were determined as described previously.49
RNA fluorescent in-situ hybridization (FISH)
RNA FISH was performed using the RNAscope® Fluorescent Multiplexed reagent kit (Advanced Cell Diagnostics, USA) as per the manufacturer’s instructions. The probe set for LASI transcript consisted of 20 dual probes targeting different segments within the whole transcript (Advanced Cell Diagnostics). Deparaffinized sections were pretreated with hydrogen peroxide solution and targets were unmasked by steaming with RNAscope® Target Retrieval buffer and with RNAScope® protease III (for cultured cells) and protease plus (for tissues) solutions for 10 min respectively at RT. Probes were hybridized for 2 h at 40 °C using a HyBEZ® oven and the signal was amplified by tandem incubation with Amp-1, Amp-2, Amp-3 and HRP-tagged probe for 30, 15, 30, and 15 min, respectively, at 40 °C using a HyBEZ® oven with thorough washing in-between. Probes were detected using Tyramide signal amplification (TSA) reaction using an Alexa-flour labeled TSA kit (PerkinElmer) as per manufacturer’s instructions. FISH sections were then either processed for immunostaining as described above or were directly stained with DAPI-containing Fluormount-G. Multifluorescent images were captured with BZX700 Microscopy system (Keyence) and analyzed by NIH ImageJ software.
RNA FISH expression was quantified by RNAscope® data analysis suite (Advanced Cell Diagnostics). Probe signals (dots)/cell were counted and allotted to separate bins as follows: Bin 0 (0 Dots/Cell); Bin 1 (1–3 Dots/Cell); Bin 2 (4–9 Dots/Cell); Bin 3 (10–15 Dots/Cell); Bin 4 (>15 Dots/Cell). Then, the Histo-score (H-Score) was calculated as follows: H-Score = Sum of each (bin number × percentage of cells per bin) i.e., H-score = (0 × % cells in Bin 0) + (1 × % cells in Bin 1) + (2 × % cells in Bin 2) + (3 × % cells in Bin 3) + (4 × % cells in Bin 4). Final scores derived by this metric have a range between 0 and 400.
Silencing RNA (siRNA) transfection
Primary HAECs cultured on plastic at 50% confluency were transfected with siRNA specific to LASI (siLASI, custom made by IDT, Inc.) or control siRNAs (siCTRL) using X2 (Mirus biotechnologies) transfection reagent (custom made by IDT technologies Inc.) as per manufacturer’s instructions. Twenty-four-hour post-transfection, cells were treated with 100 ng/ml LPS or 10 mg/ml IL-13, and 24 h later cells were analyzed.
Statistical analysis
Grouped results were expressed as means ± SEM. Data were analyzed using GraphPad Prism Software (GraphPad Software Inc.) using one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test or as indicated. Grouped results were analyzed using two-way ANOVA. When significant main effects were detected (P < 0.05), Fisher’s least significant difference and student’s t test was used to determine differences between groups.
References
Bateman, E. D. et al. “Global strategy for asthma management and prevention: GINA executive summary.”. Eur. Respir. J. 31, 143–178 (2008). Eur Respir J 2018; 51(2).
Tung, H. Y. et al. Advances and Evolving Concepts in Allergic Asthma. Semin Respir. Crit. Care Med. 39, 64–81 (2018).
Gon, Y. & Hashimoto, S. Role of airway epithelial barrier dysfunction in pathogenesis of asthma. Allergol. Int. 67, 12–17 (2018).
Lambrecht, B. N. & Hammad, H. The airway epithelium in asthma. Nat. Med. 18, 684–692 (2012).
Fahy, J. V. & Dickey, B. F. Airway mucus function and dysfunction. N. Engl. J. Med. 363, 2233–2247 (2010).
Mall, M. A., Danahay, H. & Boucher, R. C. Emerging Concepts and Therapies for Mucoobstructive Lung Disease. Ann. Am. Thorac. Soc. 15, S216–S226 (2018).
Benam, K. H., Vladar, E. K., Janssen, W. J. & Evans, C. M. Mucociliary defense: emerging cellular, molecular, and animal models. Ann. Am. Thorac. Soc. 15, S210–S215 (2018).
Bonser, L. R. & Erle, D. J. Airway Mucus and Asthma: the Role of MUC5AC and MUC5B. J. Clin. Med. 6, 112 (2017).
Saco, T. V., Breitzig, M. T., Lockey, R. F. & Kolliputi, N. Epigenetics of Mucus Hypersecretion in Chronic Respiratory Diseases. Am. J. Respir. Cell Mol. Biol. 58, 299–309 (2018).
Netea, M. G. et al. Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098 (2016).
Netea, M. G. & Joosten, L. A. B. Trained Immunity and Local Innate Immune Memory in the Lung. Cell 175, 1463–1465 (2018).
Whitsett, J. A. & Alenghat, T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16, 27–35 (2015).
Weitnauer, M., Mijosek, V. & Dalpke, A. H. Control of local immunity by airway epithelial cells. Mucosal Immunol. 9, 287–298 (2016).
Hartl, D. et al. Innate immunity of the lung: from basic mechanisms to translational medicine. J. Innate Immun. 10, 487–501 (2018).
Bigot, J. et al. Respiratory epithelial cells can remember infection: a proof of concept study. J. Infect. Dis. 221, 1000–1005 (2019).
Chand, H. S., Mebratu, Y. A., Montera, M. & Tesfaigzi, Y. T cells suppress memory-dependent rapid mucous cell metaplasia in mouse airways. Respir. Res. 17, 132 (2016).
Fernandes, J. C. R., Acuna, S. M., Aoki, J. I., Floeter-Winter, L. M. & Muxel, S. M. Long Non-Coding RNAs in the regulation of gene expression: physiology and disease. Non-coding RNA 5, 17 (2019).
Yao, R. W., Wang, Y. & Chen, L. L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551 (2019).
Devadoss, D. et al. Long Noncoding Transcriptome in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 61, 678–688 (2019).
Zhu, Y., Mao, D., Gao, W., Han, G. & Hu, H. Analysis of lncRNA Expression in Patients With Eosinophilic and Neutrophilic Asthma Focusing on LNC_000127. Front. Genet. 10, 141 (2019).
Austin, P. J. et al. Transcriptional profiling identifies the long noncoding RNA plasmacytoma variant translocation (PVT1) as a novel regulator of the asthmatic phenotype in human airway smooth muscle. J. Allergy Clin. Immunol. 139, 780–789 (2017).
Park, K. S. et al. SPDEF regulates goblet cell hyperplasia in the airway epithelium. J. Clin. Investig. 117, 978–988 (2007).
Lumsden, A. L. et al. ICAM-1-related long non-coding RNA: promoter analysis and expression in human retinal endothelial cells. BMC Res. Notes 11, 285 (2018).
Janson, C. et al. Circulating adhesion molecules in allergic and non-allergic asthma. Respir. Med. 99, 45–51 (2005).
Chand, H. S., Harris, J. F. & Tesfaigzi, Y. IL-13 in LPS-Induced Inflammation Causes Bcl-2 Expression to Sustain Hyperplastic Mucous cells. Sci. Rep. 8, 436 (2018).
Yu, H., Li, Q., Kolosov, V. P., Perelman, J. M. & Zhou, X. Interleukin-13 induces mucin 5AC production involving STAT6/SPDEF in human airway epithelial cells. Cell Commun. Adhes. 17, 83–92 (2010).
Kirjavainen, P. V. et al. Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat. Med. 25, 1089–1095 (2019).
Stein, M. M. et al. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N. Engl. J. Med. 375, 411–421 (2016).
Schuijs, M. J. et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 349, 1106–1110 (2015).
Zheng, M. et al. Long Noncoding RNA COPDA1 Promotes Airway Smooth Muscle Cell Proliferation in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 61, 584–596 (2019).
Li, Z. et al. lncRNA Malat1 modulates the maturation process, cytokine secretion and apoptosis in airway epithelial cell-conditioned dendritic cells. Exp. Ther. Med. 16, 3951–3958. (2018).
Zhu, Y. J., Mao, D., Gao, W. & Hu, H. Peripheral whole blood lncRNA expression analysis in patients with eosinophilic asthma. Med. (Baltim.) 97, e9817 (2018).
Lin, J., Feng, X., Zhang, J. & Tong, Z. Long noncoding RNA TUG1 promotes airway smooth muscle cells proliferation and migration via sponging miR-590-5p/FGF1 in asthma. Am. J. Transl. Res. 11, 3159–3166 (2019).
Fan, M., Xu, J., Xiao, Q., Chen, F. & Han, X. Long non-coding RNA TCF7 contributes to the growth and migration of airway smooth muscle cells in asthma through targeting TIMMDC1/Akt axis. Biochem. Biophys. Res. Commun. 508, 749–755 (2019).
Cabili, M. N. et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20 (2015).
Long, Y., Wang, X., Youmans, D. T. & Cech, T. R. How do lncRNAs regulate transcription? Sci. Adv. 3, eaao2110 (2017).
Mitchell, P. D. & O’Byrne, P. M. Epithelial-Derived Cytokines in Asthma. Chest 151, 1338–1344 (2017).
Khair, O. A., Devalia, J. L., Abdelaziz, M. M., Sapsford, R. J. & Davies, R. J. Effect of erythromycin on Haemophilus influenzae endotoxin-induced release of IL-6, IL-8 and sICAM-1 by cultured human bronchial epithelial cells. Eur. Respir. J. 8, 1451–1457 (1995).
Jevnikar, Z. et al. Epithelial IL-6 trans-signaling defines a new asthma phenotype with increased airway inflammation. J. Allergy Clin. Immunol. 143, 577–590 (2019).
Hadjicharalambous, M. R. et al. Long Non-coding RNAs Are Central Regulators of the IL-1beta-Induced Inflammatory Response in Normal and Idiopathic Pulmonary Lung Fibroblasts. Front. Immunol. 9, 2906 (2018).
Perry, M. M. et al. Role of non-coding RNAs in maintaining primary airway smooth muscle cells. Respir. Res. 15, 58 (2014).
Qian, X., Shi, S. & Zhang, G. Long non-coding RNA antisense non-coding RNA in the INK4 locus expression correlates with increased disease risk, severity, and inflammation of allergic rhinitis. Med. (Baltim.) 98, e15247 (2019).
Sailland, J. et al. Role of Smad3 and p38 Signalling in Cigarette Smoke-induced CFTR and BK dysfunction in Primary Human Bronchial Airway Epithelial Cells. Sci. Rep. 7, 10506 (2017).
Chung, S. et al. Electronic Cigarette Vapor with Nicotine Causes Airway Mucociliary Dysfunction Preferentially via TRPA1 Receptors. Am. J. Respir. Crit. Care Med. 200, 1134–1145 (2019).
Sweeney, T. E. et al. Unsupervised Analysis of Transcriptomics in Bacterial Sepsis Across Multiple Datasets Reveals Three Robust Clusters. Crit. Care Med. 46, 915–925 (2018).
Tsalik, E. L. et al. Renal systems biology of patients with systemic inflammatory response syndrome. Kidney Int. 88, 804–814 (2015).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Tsalik, E. L. et al. An integrated transcriptome and expressed variant analysis of sepsis survival and death. Genome Med. 6, 111 (2014).
Chand, H. S., Mebratu, Y. A., Kuehl, P. J. & Tesfaigzi, Y. Blocking Bcl-2 resolves IL-13-mediated mucous cell hyperplasia in a Bik-dependent manner. J. Allergy Clin. Immunol. 140, 1456–1459. e1459 (2017).
Chand, H. S. et al. Cigarette smoke and HIV synergistically affect lung pathology in cynomolgus macaques. J. Clin. Investig. 128, 5428–5433 (2018).
Acknowledgements
UNC CF Tissue Procurement and Cell Culture Core is supported by NIH P30DK065988 and CF Foundation BOUCHE15R0 grants. The authors appreciate the technical support provided by Christopher Long and Ruben Castro. Authors acknowledge the funding support by NIH R21 AI144374, R21 AI117560, and 1UL1TR001417; American Lung Association RG306208; and the FIU HWCOM and ORED Start-Up Funds (to H.S.C.). NIH R01HL133240, R01HL139365, FAMRI CIA160011, and the James and Esther King Florida Biomedical Research Program 5JK02 (to M.S.).
Author information
Authors and Affiliations
Contributions
D.D. conducted the experiments, analyzed the data, and wrote the paper; G.D., M.M., D.H., and S.S.H. conducted the experiments and analyzed the data; N.B. and M.S. helped design experiments and provided cell culture support; G.B. and R.J. help design the studies, analyzed the data, and wrote the paper; H.S.C. conceptualized and designed the studies, analyzed the data, and wrote the paper. All authors reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Devadoss, D., Daly, G., Manevski, M. et al. A long noncoding RNA antisense to ICAM-1 is involved in allergic asthma associated hyperreactive response of airway epithelial cells. Mucosal Immunol 14, 630–639 (2021). https://doi.org/10.1038/s41385-020-00352-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-020-00352-9
This article is cited by
-
Indoor-air purification by photoelectrochemical oxidation mitigates allergic airway responses to aerosolized cat dander in a murine model
Scientific Reports (2023)
-
ICAM-1 related long noncoding RNA is associated with progression of IgA nephropathy and fibrotic changes in proximal tubular cells
Scientific Reports (2022)