Abstract
Priming at the site of natural infection typically elicits a protective T cell response against subsequent pathogen encounter. Here, we report the identification of a novel fungal antigen that we harnessed for mucosal vaccination and tetramer generation to test whether we can elicit protective, antigen-specific tissue-resident memory (Trm) CD4+ T cells in the lung parenchyma. In contrast to expectations, CD69+, CXCR3+, CD103− Trm cells failed to protect against a lethal pulmonary fungal infection. Surprisingly, systemic vaccination induced a population of tetramer+ CD4+ T cells enriched within the pulmonary vasculature, and expressing CXCR3 and CX3CR1, that migrated to the lung tissue upon challenge and efficiently protected mice against infection. Mucosal vaccine priming of Trm may not reliably protect against mucosal pathogens.
Similar content being viewed by others
Introduction
Host defense against respiratory pathogens is thought to be mediated by lung tissue-resident memory (Trm) T cells that occupy the parenchyma without recirculating.1 Because Trm cells occupy frontline mucosal sites of infection, they are anatomically positioned to respond immediately.2 Indeed, many different microbial pathogen challenge models have shown that Trm cells mediate protection in the lung and other organs.3,4,5,6,7,8,9,10,11,12,13 The expression of the phenotypic markers CD69, CXCR3, and CD1032,3,14 allow newly primed T cells to seed nonlymphoid tissue during the effector phase and reside permanently in situ.15,16 Most of the work to date investigating Trm and their phenotypic markers pertains to CD8+ T cells.3,4,5,6,7,8,9,10,11,12,13
Vaccination at the mucosa is often advocated as the ideal way to elicit T-cell immunity to respiratory infections.17,18 Mucosal vaccination can induce long-lasting antigen-specific memory immune responses in both the systemic and mucosal compartments.18 The current licensed live-attenuated vaccines for humans against rotavirus, poliovirus, Salmonella typhi, and cholera are delivered orally, and against influenza virus (FluMist) it is applied intranasally.19 However, safety concerns persist with live-attenuated vaccines and subunit vaccines are needed. When developing subunit mucosal vaccines, identifying protective lymphocyte antigens and adjuvants that drive a proinflammatory immune response is essential.
We recently described Blastomyces endoglucanase-2 (Bl-Eng2), a novel fungal ligand for dectin-2 that induces the production of IL-1β and IL-6 by dendritic cells and acts as an adjuvant to promote differentiation of CD4+ T cells into antifungal Th17 cells.20,21 In the current study, we discovered that Bl-Eng2 also harbors a CD4+ T cell epitope(s) that can be harnessed for subunit vaccination. Therefore, we sought to investigate whether mucosal immunization with Bl-Eng2 induces the development of antigen-specific Trm cells in the lung to protect mice against infection with inhaled fungi. We found that intranasal vaccination with Bl-Eng2 induced the generation of tetramer+, CD69+, CXCR3+, CD103− Trm cells in lung tissue. However, in contrast to our expectations and predications, we found that mucosal vaccination and CD4+ Trm cells failed to protect against respiratory challenge with B. dermatitidis, whereas systemic vaccination did confer protection. Bl-Eng2-specific T cells from subcutaneously (SC) vaccinated mice migrated from the secondary lymphoid organs (SLOs) and the lung vasculature to the parenchyma upon challenge and reduced lung CFU. Migration of protective T cells was correlated with increased expression of the chemokine receptors CX3CR1 and CXCR3 on tetramer+ cells within the lung vasculature and parenchyma. Thus, vaccine-induced protection against a lethal pulmonary fungal infection was not mediated by lung CD4+ Trm cells following mucosal immunization, but rather by a population of migratory CD4+ T cells after systemic vaccination.
Results
Identification of Bl-Eng2 as a protective antigen and generation of a tetramer to track and enumerate Ag-specific CD4+ T cells in vaccine immunity
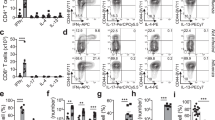
We recently described Bl-Eng2 as a yeast ligand for dectin-2.21 Bl-Eng2 is more potent than other reported dectin-2 ligands in stimulating accessory cells, and exerts adjuvant activity by eliciting IL-1β and IL-6 from dendritic cells,21 cytokines that are instrumental in driving differentiation of CD4+ T cells into Th17 cells.22 Our initial work reported that Bl-Eng2 served as an adjuvant, augmenting calnexin-induced vaccine immunity against experimental blastomycosis.21 Here, we asked if Bl-Eng2 could act as both an adjuvant and antigen. Bl-Eng2 formulated in incomplete Freund’s adjuvant (IFA) and delivered SC in vaccinated mice (IFA acts as a depot to simply retain Bl-Eng2 in tissue) carried sufficient activity as an adjuvant and antigen to confer resistance against experimental blastomycosis. Bl-Eng2 in IFA reduced lung colony forming units (CFU) of yeast by >3 logs, compared with controls that received IFA vehicle alone (Fig. 1a). Thus, Bl-Eng2 is both an adjuvant and a protective antigen that significantly enhances resistance and prolongs survival (data not shown) against infection.
a Lung CFU in SC vaccinated and control mice day 11 post infection. Data are representative of three independent experiments (n = 5–10 mice/ group). *p < 0.05 vs. IFA control mice. Number indicates n-fold difference vs. IFA control group. b Synthetic peptides (nine amino acid [aa] core plus 2 flanking aa on either side) used for ex vivo stimulation of Bl-Eng2 primed T cells harvested from splenocytes of SC-vaccinated mice. c IFN-γ production in cell culture supernatants measured by ELISA. *p < 0.05 vs. all other groups. d At day 4 post infection, CD4+ (top row) and CD8+ (bottom row) T cells from the lung were labeled with tetramer. Numbers indicate the percentage of parent gate.
We sought to develop tools to resolve endogenous antigen-specific T cell immune responses after mucosal vaccination. We mapped the Bl-Eng2 peptide epitope recognized by CD4+ T cells and generated class II MHC tetramers using methods described.23,24 We first analyzed Bl-Eng2 for MHC class II binding sequences. Of five predicted peptides from Bl-Eng2, one 13-mer (AFFDGPDPSNAYV; peptide #1) that begins at residue 35 significantly activated CD4+ T cells from splenocytes of mice vaccinated with Bl-Eng2 (Fig. 1b, c). Other peptides and stimuli (except for Bl-Eng2 protein) elicited little or no response.
Using this peptide, we created an MHC class II tetramer that revealed expansion and recruitment of primed Bl-Eng2 antigen-specific CD4+ T cells into the lungs of vaccinated mice (Fig. 1d). Four days after challenge, ≈10% of CD4+ T cells recruited to lung were tetramer+ CD44+. The tetramer was specific. Few CD8+ T cells bound tetramer.
Vaccination at the respiratory mucosa elicits strong T-cell immunity but fails to protect against inhaled fungi
Vaccination at the mucosa is viewed as the ideal strategy to foster resistance against a mucosal pathogen. For example, a recent study found that intranasal (IN) influenza vaccination induced resistance against experimental infection.3 We therefore formulated Bl-Eng2 in glucan–chitin particles (GCPs), which we have reported previously,24 and vaccinated mice IN three times, spaced 2 weeks apart; in parallel, Bl-Eng2 in GCPs was given SC (Fig. 2a). IN vaccination efficiently elicited Bl-Eng2 specific CD4+ T cells in the lung and spleen (Fig. 2b, d). However, the number of tetramer+ cells was threefold higher in the lung and eightfold higher in the spleen of SC-vaccinated mice compared with IN-vaccinated mice. After challenge, the number of tetramer+ cells recalled to the lungs was also twofold higher in SC-vaccinated mice than in IN-vaccinated mice (Fig. 2c). Nevertheless, >105 tetramer+ CD44+ CD4+ T cells were recruited to the lung parenchyma for both routes. In comparison, we previously reported ≈100 tetramer+ CD4+ T cells recalled to the lung of mice successfully vaccinated against infection with calnexin and complete Freund’s adjuvant (CFA);24 a number that is orders of magnitude lower than with Bl-Eng2.
a Mice received Bl-Eng2 in GCP either SC or IN three times, 2 weeks apart. CD4+ tetramer+ T cells were enumerated in the lung before (b) and after (c) challenge (day 4 post-infection). Anti-CD45 mAb was injected i.v. 5 min before euthanizing mice to stain lung vascular cells; plots in the second row show total tetramer+ cells distributed in the parenchyma and lung vasculature. Numbers in plots indicate either mean ± SEM or percent of parent gate. Contour plots show combined data from five independent experiments (n = 25–30 mice/group). d Cells from the spleen of unchallenged mice were pulled down with Bl-Eng2 tetramer and enumerated. Numbers in plots indicate mean ± SEM (n = 4 mice/group). *p < 0.05 for SC vs. IN vaccine groups. e Resistance to infection as determined by lung CFU. Mouse serum albumin (MSA) on GCPs served as a control (n = 10 mice/group). CFU graph shows geometric mean with standard deviation. *p < 0.05 vs. control mice.
Remarkably, we found no evidence of resistance to infection in IN-vaccinated mice with Bl-Eng2, whereas SC-vaccinated mice did acquire resistance (Fig. 2e). We considered that the adjuvant GCP might be the source of the mucosal vaccine failure. Thus, we next vaccinated mice IN with another adjuvant, Adjuplex, which is a biodegradable carbomer homopolymer and submicron-sized liposomes [nanoliposomes] derived from purified soy lecithin. Adjuplex-based vaccination induced resistance against experimental infection with influenza.3 Bl-Eng2 in Adjuplex given IN was well tolerated and elicited strong immune responses upon vaccination revealing >10,000 tetramer+ cells in the lung before and after challenge (SFig. 2A–D). Yet, Bl-Eng2 given IN with Adjuplex again failed to protect mice against infection, while Bl-Eng2 in CFA given SC did protect mice (SFig. 2E). Thus, we concluded that the route of vaccination and not the mucosal adjuvant likely accounts for the observed resistance phenotype. For our subsequent experiments, we used GCP as the vaccine adjuvant to provide consistency in vaccine formulation between the routes of vaccination.
Mucosal vaccination induces increased Treg cell numbers and reduced IFN-γ responses by tetramer+ T cells
Both IFN-γ and IL-17 CD4+ T cells confer antifungal vaccine immunity.22,25,26,27 We therefore analyzed transcription factor expression and the frequencies and numbers of cytokine-producing, Bl-Eng2-specific T cells in the lung at day 4 post infection to see if there was a difference between IN- and SC-vaccinated mice. We found that SC-vaccinated mice showed a trend towards increased T-bet expression, while IN-vaccinated mice had significantly higher MFI for ROR-γt (Fig. 3a). Likewise, the frequencies of IFN-γ+ tetramer+ T cells were increased in SC-vaccinated mice, whereas the frequencies of IL-17 producing T cells were increased in IN-vaccinated mice (Fig. 3b). Consequently, the number of Th1 cells in the lung is increased in SC-vaccinated mice, whereas the number of Th17 cells is comparable between the two vaccine groups (Fig. 3c). The expression of GATA3 (Fig. 3a) and the frequencies of IL-5/IL-13+ tetramer+ cells also were similar in the two groups (Fig. 3b). However the number of Th2 cells was higher in SC-vaccinated mice (Fig. 3c), mainly because more tetramer+ T cells were found in the lung of SC-vaccinated mice (Fig. 2b). Similar proportions and numbers of Th1/Th17 cells in the lung and spleen were found in vaccinated mice before challenge (SFig. 3).
Mice received Bl-Eng2 in GCP either SC or IN three times, 2 weeks apart. a Two weeks following the last boost, mice were challenged and lungs were harvested at day 4 post infection and stained for transcription factors T-bet, ROR-γt, and GATA3. Histograms show expression of each transcription factor within the designated tetramer+ cell populations. Adjacent bar graphs show corresponding MFI values for individual mice. b Dot plots display cytokine producing or foxp3+ lung tetramer+ cells at day 4 post infection; cells in plots for IFN-γ, IL-17, and IL-5 and IL-13 were stimulated ex vivo with Bl-Eng2 peptide for 5 h; foxp3 staining was performed before stimulation on a separate lung aliquot. Numbers in plot represent percent of parent gate. c Absolute numbers of cytokine producing T helper and regulatory T (foxp3+) cells. Data are from one representative experiment of two performed (n = 5 mice/group). *p < 0.05 for SC vs. IN vaccine groups. d Lung CFU of SC-vaccinated mice treated with cytokine neutralizing antibody. SC-vaccinated mice with Bl-Eng2 in GCP were given 100 µg i.v. of rat IgG or neutralizing cytokine antibody on the day of challenge and every other day thereafter until harvest at day 7 post-infection. Data shown are a combination of two independent experiments (n = 10 mice/group). *p < 0.05 for rat IgG vs. indicated treatment groups.
Both IFN-γ and IL-17 have been linked to vaccine resistance.22,25,26,27 We therefore sought to functionally test the role of these two cytokines and neutralized them in SC-vaccinated mice during the expression phase (after infection). Neutralization of IFN-γ or IL-17 alone or together resulted in elevated fungal burdens compared with rat IgG treated controls (Fig. 3d). These data indicate that both cytokines contribute to Bl-Eng2-induced immunity in SC-vaccinated mice.
To investigate why Th17 cells fail to mediate vaccine protection in IN-vaccinated mice, we enumerated the proportion and number of regulatory T (Treg) cells by staining tetramer+ cells for the transcription factor Foxp3. The frequency and numbers of foxp3+ tetramer+ cells were significantly higher in IN-vaccinated mice (Fig. 3b, c). Interestingly, Treg cells outnumbered Th1 cells in IN-vaccinated mice (Fig. 3c). Thus, it is possible that Treg cells suppress protective Th17 cells in IN-vaccinated mice.
IN vaccination induces CD69+ Trm cells, whereas SC vaccination elicits CXCR3+ and CX3CR1+ lung tetramer+ T cells
In a model of Mycobacterium tuberculosis infection,9 the authors used the phenotypic markers CD69, CD103, CXCR3, CX3CR1, and KLRG1 to distinguish protective parenchymal Th1 cells from nonprotective, vascular CD4+ T cells. In their experimental model, the phenotype of lung vascular cells was CD69−, CXCR3−, CX3CR1+, and KLRG1+, whereas that of parenchymal cells was CD69+, CXCR3+, CX3CR1−, KLRG1−. We assessed expression of the Trm markers CXCR3, CD69, CD103,2,28,29,30 the vascular marker CX3CR1, and the terminal differentiation marker KLRG1.31 We phenotypically analyzed tetramer+ T cells in the lung parenchyma and vasculature before and after challenge. We found significant differences in CD69, CXCR3, CX3CR1, and KLRG1 expression by tetramer+ T cells from IN-vaccinated mice compared with SC-vaccinated mice (Fig. 4a, b). In the lung parenchyma, CD69 expression was increased whereas CXCR3 expression was decreased in the IN-vaccinated compared with SC-vaccinated mice. Interestingly, CXCR3 expression was lost in the cytokine-producing T cells after challenge (SFig. 4A, B). In the vasculature, the expression of CX3CR1 and dual expression of CX3CR1 KLRG1 was increased in SC-vaccinated mice compared with IN-vaccinated mice (Fig. 4a, b). We did not detect significant expression of CD103 by lung tetramer+ cells in either compartment or route of vaccination (Fig. 4a, b).
Mice were vaccinated with Bl-Eng2 in GCP either SC or IN. Two weeks after the last boost, lungs were harvested (a) prior to challenge and (b) at day 4 post infection. Total lung tetramer+ T cells were separated into parenchymal and vasculature cells based on i.v. anti-CD45 mAb staining and were analyzed for the phenotypic markers CD69, CXCR3, CX3CR1, CD103, and KLRG1. Numbers in the dot plots and graphs below indicate percent of the parent gate, either tetramer+CD45 i.v.− (parenchyma) or tetramer+CD45 i.v.+ (vasculature). Data are a combination of three independent experiments (n = 15 mice/group). *p < 0.05 for SC vs. IN vaccine groups. Heatmap tSNE plots for lung cells before (c) and after (d) challenge from one representative experiment of three performed (n = 5 mice/group). In FlowJo, total tetramer+ cells from individual mice were concatenated into a single FSC file for SC and IN groups before and after challenge. tSNE parameters (iterations: 1000; perplexity: 30) were computed for this concatenated file including the markers CD45 i.v., CD69, CXCR3, CD103, CX3CR1, and KLRG1. Heatmap tSNEs show individual vaccine and challenge groups plotted based on computed tSNE parameters 1 and 2; these parameters allow visualization of the six listed markers in two dimensional space by clustering cells with similar expression profiles together. The scale bar reflects the relative expression of the labeled marker; red and yellow indicate high expression compared to blue for low expression.
Hierarchical gating used in flow cytometry is limited by simultaneous viewing of two parameters. This restriction increases the opportunity for user biases in the gating analysis. To take an impartial approach to our traditional phenotypic analysis, we used the t-distributed stochastic neighbor embedding (tSNE) algorithm to evaluate marker expression on tetramer+ T cells. The tSNE algorithm visualizes complex data obtained from cell staining with multiple markers by clustering cells with similar expression patterns. The ability to visualize several markers in one plot provides a holistic view of a whole cell population inaccessible by classical flow analysis. The two routes of vaccination lead to different clustering patterns for CD69, CXCR3, and CX3CR1 among the tetramer+ cells (note circled populations in Fig. 4c, d). Before challenge, IN-vaccinated mice had more tetramer+ T cells with high levels of CD69 than did SC-vaccinated mice. After challenge, however, the two vaccine groups were similar in this CD69 phenotype. On the other hand, SC-vaccinated mice had more tetramer+ cells with high levels of CXCR3 within the lung parenchyma and vasculature both before and after challenge, when compared with IN-vaccinated mice (note circled populations in Fig. 4c, d and SFig. 4C, D). SC-vaccinated mice also had more tetramer+ cells expressing CX3CR1 and KLRG1 within the lung vasculature both before and after challenge, whereas these populations were absent in the IN-vaccinated mice (SFig. 4C, D).
In summary, delivery of Bl-Eng2 by the respiratory route elicited CD69+ Trm cells, whereas SC vaccination induced the generation of tetramer+ T cells that expressed the chemokine receptors CXCR3 and CX3CR1 and the terminal differentiation marker KLRG1. These chemokine receptors may facilitate migration of primed T cells from the pulmonary vasculature and SLO to the lung parenchyma.
Migration of systemic tetramer+ T cells from the SLO to the lung is delayed and reduced in IN-vaccinated mice
Since the total number of tetramer+ T cells (Fig. 2) in the lung vasculature is higher in SC-vaccinated mice than in IN-vaccinated mice, we sought to assess whether the vasculature is the final destination of recalled, tetramer+ CD4+ T cells or a transient location on their route of migration to the lung tissue. To answer this question, we studied the kinetics of T cell location before and after challenge. We analyzed mice vaccinated by each route for the frequency and number of tetramer+ cells in different anatomical locations of the lung before infection and at serial time points post infection. Twenty-four hours prior to harvest of lung T cells, mice were given an initial anti-CD45 fluorescent mAb i.v. to mark T cells that were accessible in the blood stream and lung blood vasculature (Fig. 5a). In this maneuver, T cells in the lung parenchyma are protected against i.v. staining. A second CD45 mAb conjugated to a different fluorochrome was injected i.v. 5 min before mice were sacrificed.
a Mice were vaccinated with Bl-Eng2 in GCP either SC or IN three times, 2 weeks apart. Two weeks after the last boost, mice were challenged with B. dermatitidis yeast. Twenty-four hours before the harvest, and at serial time points post infection, an initial anti-CD45 mAb (BV412) was given to mice by i.v. injection. On the day of harvest 5 min before euthanizing mice, a second anti-CD45 mAb conjugated to a different fluorochrome (Alexa 488) was also given i.v. Tetramer+ cells that stained for the second and initial mAb were termed vascular; those that stained for the initial but not the second mAb were termed migratory; and those that did not stain for either mAb were termed tissue. b Dot plots show the frequencies of tetramer+ cells that stained for the second vs. initial anti-CD45 mAb. The line graph shows total number of tetramer+ T cells over time. c Graphs show frequencies of tetramer+ cells found in corresponding compartments on serial days post infection. d Lung CFU at serial days post infection. e Numbers of total (left), vascular (middle) and migratory (right) tetramer+ cells positive for CXCR3+ or CX3CR1+. Data are from one representative experiment of two independent ones performed (n = 5 mice/group). *p < 0.05 vs. corresponding IN vaccine group.
Before challenge, over 70% of tetramer+ cells resided in the lung parenchyma in SC-vaccinated mice (Fig. 5b) and 18% of tetramer+ cells were marked in the vasculature (day 0). Upon challenge, at day 1 post infection, new tetramer+ cells migrated into the lung vasculature (17%) and cells previously in the vasculature migrated into the lung tissue (35%). By days 2 and 3 post infection, the frequency of tetramer+ cells entering the lung tissue from the vasculature in SC-vaccinated mice (54–66%) were significantly higher than that in IN-vaccinated mice. At day 4 post infection, migration through the blood vasculature and lung tissue diminished and most of the tetramer+ cells in the lung resided in the tissue. Migration of tetramer+ T cells into the lung tissue of IN-vaccinated mice was delayed (by 1 day) and reduced during peak migration (days 2 and 3), compared with SC-vaccinated mice, as assessed by the frequency and number of cells in the vasculature and freshly migrated into the tissue (Fig. 5b, c and SFig. 5A). Thus, tetramer+ cells elicited by the SC vaccine migrate from the pulmonary vasculature into the lung parenchyma upon fungal infection. The vasculature is thus either a depot or transient location for migrating Ag-specific T cells. The influx of migratory T cells into the lung parenchyma coincided with a reduction in lung CFU in SC-vaccinated mice, but not IN-vaccinated mice (Fig. 5d). The number and frequency of CXCR3+ and CX3CR1+ tetramer+ T cells were greater among the migratory and vascular T cells for SC-vaccinated mice than IN-vaccinated mice, suggesting these two chemokine receptors may promote their migration (Fig. 5e and SFig. 5B).
CXCR3 and CX3CR1 are dispensable for T-cell migration and vaccine-induced resistance
To investigate whether CXCR3 and CX3CR1 are functionally required for T-cell migration from the SLO to the lung upon challenge we SC-vaccinated CXCR3−/−, CX3CR1−/− mice and wild-type controls with GCP-Bl-Eng2. We confirmed the loss of chemokine receptor expression by surface staining in the corresponding KO mice (SFig. 6). At day 4 post infection, the number of tetramer+ cells that migrated to the lung, proliferated and produced Th1, Th17, and Th2 cytokines was comparable among the CXCR3−/−, CX3CR1−/− mice and wild-type controls (Fig. 6a, b and SFig. 6). Lung CFU was likewise comparable across the three groups (Fig. 6c). These data indicate that CXCR3 and CX3CR1 are dispensable for T-cell migration and Bl-Eng2 induced immunity.
C57BL6 (WT), CXCR3−/−, and CX3CR1−/− mice were SC vaccinated with Bl-Eng2 in GCP or MSA in GCP three times, 2 weeks apart. Two weeks after the last boost mice were challenged and lungs were harvested at day 4 post infection. a Dots plots show tetramer+ T cells, numbers on plot indicate percent of parent gate. Corresponding bar graphs display absolute number of tetramer+ cells for WT and KO strains. Anti-CD45 mAb was injected i.v. 5 min before euthanizing mice to stain lung vascular cells; plots in the second row show total tetramer+ cells distributed in the parenchyma and lung vasculature. b Cytokine production by tetramer+ cells after stimulation ex vivo with Bl-Eng2 peptide. Dot plots show frequency among total tetramer+ cells and bar graphs below display absolute numbers of cytokine-producing tetramer+ cells. c Lung CFU at day 4 post infection (n = 5 mice/group). *p < 0.05 for naive vs. vaccinated groups.
After recall, proliferation of lung T cells is increased in IN-vaccinated mice
Despite increased migration of tetramer+ cells from the vasculature and SLO into the lung parenchyma for SC-vaccinated mice compared with IN-vaccinated mice, the number of tetramer+ cells in the lung tissue was similar between the two routes of vaccination at days 4 post infection (Fig. 5b). We hypothesized that tetramer+ cells might proliferate more after challenge in IN-vaccinated mice than SC-vaccinated mice. To test this hypothesis, we BrdU-labeled proliferating tetramer+ T cells between the day of challenge and day 4 post infection. Indeed, tetramer+ and cytokine-producing T cells in the lung parenchyma elicited during the recall response proliferated more in the IN-vaccinated mice than in the SC-vaccinated mice (Fig. 7). Nevertheless, despite the compensatory proliferation of Trm cells, the IN-vaccinated mice were not protected against fungal infection (Fig. 2e).
Mice were vaccinated with Bl-Eng2 in GCPs either SC or IN. Two weeks after the last boost, mice were harvested prior to challenge or at day 4 post infection. Two micrograms of BrdU was given by i.p. injection to all mice for three consecutive days before harvest. a Dot plots and graphs show the frequencies of tetramer+ (a) and cytokine+ (b) cells that are BrdU+ prior to challenge. c, d Dot plots and graphs illustrate analogous data at day 4 post infection. Data are representative of two independent experiments (n = 5 mice/group). *p < 0.05 for IN vs. SC vaccine groups.
Migratory T cells are required and sufficient to mediate vaccine-induced protection
To investigate whether migratory cells are necessary and sufficient to confer Bl-Eng2 vaccine-induced immunity, we employed three complementary approaches. First, we systemically depleted CD4+ T cells in SC-vaccinated mice by i.v. injection of anti-CD4 mAb before fungal challenge. Consistent with the route of injection, we observed a >90% decrease in the number and frequency of CD44+ CD4+ T cells in the circulating blood, brachial lymph nodes, spleen, and lung (SFig. 8A, B). Systemic depletion of CD4+ T cells also resulted in fewer tetramer+ T cells in the lung after fungal challenge (SFig. 8C) and significantly increased fungal burden at day 6 post infection (Fig. 8a), as compared with rat IgG control-treated mice. On the other hand, intratracheal delivery of anti-CD4 mAb before challenge, which depleted CD4+ T cells in the lung (but not systemically) (SFig. 8A, B), did not reduce the number of lung tetramer+ T cells after fungal challenge (SFig. 8C), nor did it increase the lung CFU significantly at day 6 post infection (Fig. 8a). These data suggest that systemic T cells in the pulmonary vasculature and SLO, but not the lung Trm cells, are required to mediate protection in SC-vaccinated mice.
Mice were vaccinated SC with CFA + Bl-Eng2. a Prior to challenge, CD4+ T cells were depleted in the lung (by i.t. delivery of 20 μg anti-CD4 mAb) or systemically (by i.v. delivery of 100 μg anti-CD4 mAb). Lung CFU were determined at day 6 post infection. Data shown are a combination of three independent experiments (n = 25–30 mice/group). *p < 0.05 vs. indicated groups. b Mice were vaccinated SC with Bl-Eng2 in GCP or CFA + Bl-Eng2. Two days before challenge and daily thereafter, mice were given 25 μg of FTY720 inhibitor i.p. Lung CFU were determined at day 6 post infection. Data are a combination of two independent experiments (n = 20 mice/group). *p < 0.05 vs. nontreated control group. c Schematic of adoptive transfer of CD4+ T cells from the lung (Trm) or spleen (migratory) of SC-vaccinated wild-type mice into TCR-α knockout mice. d C57BL6 wild-type mice were vaccinated SC twice with CFA + Bl-Eng2. Two weeks after the boost, migratory CD4+ T cells were isolated by positive selection using anti-CD4 magnetic beads and lung resident T cells by purified by Percoll gradient. Cells were adoptively transferred into naive TCR-α knockout mice. As a negative control, CD4+ T cells from the spleen of naive wild-type mice were transferred. At day 11 post infection, lung CFU were determined. Data are a combination of two independent experiments (n = 8–20 mice/group). *p < 0.05 vs. no transfer control group.
In a second approach, to assess the role of T cells that migrate into the lungs, we treated SC-vaccinated mice before and after challenge with FTY720 (fingolimod). FTY720 inhibits the sphingosine-1-phosphate receptor pathway, thereby impairing the trafficking of lymphocytes from lymph nodes and spleen to tissues.32 FTY720 treatment reduced the number of circulating CD4+ CD44+ T cells in the blood (SFig. 8D) and significantly increased lung CFU at day 6 post infection (Fig. 8b). These data support our hypothesis that migratory T cells mediate vaccine immunity.
Finally, in a third approach, we harvested migratory CD4+ T cells from the spleen and resident T cells from the lung of SC-vaccinated mice and adoptively transferred these populations separately into naive TCR-α knockout mice before challenge (Fig. 8c). Transfer of migratory T cells, but not Trm cells, reduced lung CFU significantly at day 11 post infection, as compared with mice that received no transfer of CD4+ T cells (Fig. 8d). Taken together, our data suggest that migratory T cells, but not the lung resident T cells, i.e., the Trm, are required and sufficient to mediate Bl-Eng2 induced vaccine immunity.
Discussion
The prevailing view—based largely on work with CD8+ T cells—is that Trm cells are responsible for long-term vaccine memory.1,33 Moreover, there has been long-standing interest in developing mucosal vaccines, which preferentially prime Trm, against a variety of microbial pathogens.3,17,18 Our findings reveal that, contrary to current assumptions, vaccine protection against inhaled fungi (i) is elicited better in the skin than at the respiratory mucosa; (ii) resides in memory cells that are not Trm or resident in the lung, but rather migrate to the lung upon recall from the vasculature and lymphoid tissue; and (iii) follows distinct rules that are not predicted by Trm cell surface markers.
The rules for Trm cells have been developed mainly from data generated with CD8+ T cells.3,4,5,6,7,8,9,10,11,12,13 There is less consensus for CD4+ Trm cells mainly because there have been fewer studies. However, there is evidence for the importance of CD4+ Trm in the lung and nasal mucosa, where they may be critical for protecting against various microbes32,34,35,36,37,38,39,40 or promoting allergic asthma.39,40 CD4+ Trm cells can promote the clearance of B. pertussis,32,34 serve as an important first line of defense against viral infections35 and mediate protection against pneumococcal pneumonia.38
Many Trm stably express CD69, but CD69 expression may not be sufficient to infer residence.41 CD4+ Trm are anchored in the lung tissue. In mice, their persistence is mediated by specific adhesion molecules that prevent recirculation and allow migration in the tissue. The expression of CD103 help Trm dock to E-cadherin in epithelia.42,43 CD103+ Trm also express other integrins and adhesion molecules such as VLA-1 and the chemokine receptors CXCR6, CXCR3, and CCR5, which are associated with T cell recruitment to inflamed tissues.36
The inability of Trm to mediate vaccine-induced resistance after being primed at the lung mucosa in our model is unexpected and contrasts to a body of literature that demonstrates that Trm cells are the key defense against mucosal pathogens.3,4,5,6,7,8,9,10,11,12,13 In a murine model of primary M. tuberculosis infection, lung parenchymal and vascular Ag-specific CD4+ T cells were functionally and phenotypically distinct subsets.8 Parenchyma-localized T cells expressed the activation markers CD69, PD-1 and the chemokine receptor CXCR3, and were associated with resistance to infection, whereas T cells in the blood vessels that expressed CX3CR1+, KLRG1hi, and high levels of T-bet and IFN-γ were not protective.8 Thus, the phenotypic characterization in that setting of primary pulmonary infection indicated that parenchymal T cells associated with resistance were Trm, whereas nonprotective vascular T cells were highly differentiated, terminal effector cells that lacked the ability to enter the lung parenchyma. IN vaccination against Influenza also induced a protective population of CXCR3+ CD69+CD103+ Trm, whereas SC vaccination failed to elicit this Trm population and did not protect against infection.3
In contrast to the studies above, we found that IN vaccination with Bl-Eng2 induced classic Trm markers (CD69+, CXCR3low, CX3CR1−, KLRG1−) in lung parenchymal cells that were nonprotective, whereas SC vaccination induced a population of protective T cells that rapidly migrated from the vasculature and SLO to the lung upon challenge and appeared phenotypically distinct. In the parenchyma, tetramer+ cells from SC-vaccinated mice upregulated CD69 after challenge and were CXCR3high, CX3CR1−, KLRG1−, whereas in the vasculature the majority of the migratory T cells were CD69−, and a proportion of them were CXCR3+, CX3CR1+, and KLRG1+. Thus, chemokine receptors that are needed to seed Trm into the lung tissue may also promote the recruitment of migratory T cells from the vasculature, SLO and other sites to the lung upon challenge. Hence, there could be functional overlap of chemokine receptors at different stages of the immune response. SC vaccination of CXCR3−/−, CX3CR1−/− mice did not reveal a deficit in T-cell migration of primed T cells from the SLO to the lung upon challenge, indicating that these chemokine receptors are dispensable. However, we cannot rule out the possibility that CXCR3 and CX3CR1 and other chemokine receptors each have a cumulative effect on T-cell migration to the lung. During TB infection, Th1 cell migration into the lung was shown to be mediated by coexpression of multiple chemokine receptors, each with a relatively small contribution.44 In that study, individual chemokine receptor contributions could not be revealed at a single time point using gene KO mice. Competitive adoptive transfer approaches were required to reveal a partial, nonredundant role of individual chemokine receptors for T cell entry into the lung.
In our study, Trm cells either produced IL-17 or expressed foxp3 indicating that the latter subset were Treg, whereas migratory T cells were predominantly Th1 cells. We have previously reported that both Th17 and Th1 cells contribute to vaccine-induced protection against fungi.45,46 Here, we show that migratory T cells from SC-vaccinated mice mediate protection by producing IFN-γ and IL-17. Thus, it is unexpected that mucosal Trm producing IL-17 in IN-vaccinated mice were nonprotective. It is possible that regulatory Trm cells, which are typically not associated with host resistance to fungal infection, dampen the protective effects of Th17 cells.47,48 It has been shown, that zymosan (predominantly composed of β1,3 glucan) can induce antiinflammatory effects on T-cell differentiation and promote the generation of Treg cells.49,50 Thus, the capacity of intranasally delivered β-glucan to promote lung regulatory Trm cells may have contributed to the failure to induce protection. Nevertheless, we found that Bl-Eng2 administered IN in an alternative adjuvant, Adjuplex, likewise failed to induce protection.
Our results suggest that the vaccine route is critical for functional attributes of primed, protective T cells. Even though SC-vaccinated mice harbored a population of antigen-specific CD4+ Trm cells in the lung before challenge, these primed T cells failed to mediate resistance, in contrast to migratory cells. In addition, IN-vaccinated mice harbored a population of tetramer+ T cells in the draining lymph node and spleen that migrated in a delayed and reduced fashion to the lung upon challenge. Perhaps these cells arrived too late to curtail the growing infection, or were functionally different from migratory T cells elicited by SC vaccination. Our findings imply an important functional difference according to the anatomical compartment of accessory cells involved in antigen delivery, presentation, and priming of naive T cells. We surmise that the population of antigen presenting cells, the inflammatory milieu and the location of priming are together instrumental in generating T cells endowed with the proper migratory and protective attributes. It is also possible that secondary expansion and cytokine production of primed T cells and cross talk between the lung-associated (mediastinal) lymph node and the lung following challenge are also distinct between IN- and SC-vaccinated mice. Ongoing investigations will distinguish between these possibilities.
Materials and methods
Mice, vaccination, and fungal infection
C57BL/6 mice were obtained from Jackson Laboratory and bred at our facility. Mice were 7–8 weeks old at the time of initiation of these experiments. Mice were housed and cared for as per guidelines of the University of Wisconsin Animal Care Committee who approved all aspects of this work.
Ten micrograms of Bl-Eng2 was loaded into GCPs and mice were vaccinated SC or IN three times, 2 weeks apart as described,51 unless otherwise indicated. In some experiments, mice were vaccinated twice either SC with 10 µg Bl-Eng2 formulated in CFA or IFA, or IN with 10 µg Bl-Eng2 in 10% Adjuplex (Empirion, LLC, Columbus OH), 2 weeks apart. Two weeks after the boost, mice were challenged intratracheally (i.t.) with 2 × 104 wild-type virulent B. dermatitidis yeast (ATCC strain 26199) and analyzed for lung T cell responses at day 4 post infection, and lung CFU at day 4 or ~2 weeks post infection. Fungal burdens were measured from homogenized tissue by serial dilutions on brain heart infusion agar plates with penicillin and streptomycin.
Intravascular staining, T-cell stimulation, and flow cytometry
Mice were injected intravenously (i.v.) with 2 µg fluorochrome-labeled anti-CD45 Ab, and 5 min later lungs and SLOs were harvested. Lungs were dissociated in Miltenyi MACS tubes and digested with collagenase (1 mg/mL) and DNase (1 µg/mL) for 25 min at 37 °C. Digested lungs were resuspended in 5 mL of 40% percoll; 3 mL of 66% percoll was underlaid (GE healthcare 17–0891–01). Samples were spun for 20 min at 2000 rpm at room temperature. Lymphocytes were then harvested from the buffy coat layer and resuspended in complete RPMI (10% FBS, 1% penicillin and streptomycin). For ex vivo T-cell stimulations, cells were incubated at 37 °C for 5 h with 5 μM Bl-Eng2 peptide #1 and 1 μg anti-mouse CD28 (BD 553294); after 1 h of incubation, BD GolgiStop™ (BD 554724) was added to samples. All FACs samples were stained with invitrogen’s LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit and Fc Block for 10 min at room temperature. Cells were then stained for Bl-Eng2 tetramer for 1 h at room temperature. Finally, cells were stained for surface antigens and, if indicated, intracellular targets for 20 min at 4 °C. Transcription factor staining was performed on primed but unstimulated cells using the Foxp3 Transcription Factor Staining kit (ebioscience cat# 00–5523–00). All panels included a dump channel to decrease background in CD4+ T cells (Dump: CD11b, CD11c, NK1.1, B220, CD8). Fifty microliters of AccuCheck Counting Beads (Invitrogen PCB100) was added to all samples to determine absolute cell counts. Samples were acquired on a LSR Fortessa at the University of Wisconsin Carbone Cancer Center Flow Lab.
For cell surface analysis the following antibody cocktail was used: CD45 i.v. A488 (30-F11 Biolegend cat#103122), MHC class II tetramer-PE, CD8 PerCP-Cy5.5 (53–6.7 Biolegend cat#100734), CX3CR1 PE-Cy7 (SA011F11 Biolegend cat#149016), CXCR3 BV421 (CXCR3–173 BD cat#562937), CD69 BV510 (H1.2F3 BD cat#563030), CD44 BV650 (IM7 Biolegend cat#103049), CD103 BV786 (M290 BD cat#564322), CD11b APC (M1/70 Biolegend cat#101212), CD11c APC (N418 Biolegend cat#117310), NK1.1 APC (PK136 Biolegend cat#108710), B220 APC (RA3–62B Biolegend cat#103212), KLRG1 BUV395 (2F1 BD cat#740279), CD4 BUV737 (RM4–5 BD cat#565246). For intracellular staining the following antibodies were used: IFN-γ PE-Cy7 (XMG1.2 BD cat#557649), IL-17A BUV395 (TC11–18H10 BD cat# 565246), ROR-γt BV421 (Q31–378 BD cat#562894), T-bet A647 (4B10 Biolegend cat#653810 and 04–46 BD cat#561267 and Invitrogen 4B10 cat# 50–5825–80), Foxp3 e450 (FJK-16S Invitrogen cat#48–5773–82), GATA3 A647 (16E10A23 Biolegend cat#653809 and TWAJ Invitrogen cat#50–9966–42 and BD L50–823 cat#560078). For kinetic analysis described in Fig. 5, additional antibodies used included: CD45 i.v. BV421 (30-F11 Biolegend cat#103134) and CXCR3 BV650 (CXCR3–173 Biolegend cat#126531).
For tSNE analysis, total Bl-Eng2 tetramer+ events from individual mice were electronically concatenated into a single file; fully compensated, cleaned data collected from one representative experiment of several performed were used to generate this file. This concatenated file was not downsampled before running tSNE analysis and contained all original tetramer+ events from each mouse (88,002 total events). TSNE settings were: iterations = 1000, perplexity = 30, eta = 6161.
Tetramer pulldown
Miltenyi LS columns on a quadroMACS magnet were used to enrich tetramer+ cells from SLOs. Spleen and draining lymph nodes were harvested from SC- or IN-vaccinated GCP + Bl-Eng2 mice, mashed through 40 μm filters, and then subjected to red blood cell lysis (ACK lysis buffer 2.5 min at RT). Samples were washed with 15 mL and resuspended in cold sorter buffer (PBS with 2% FBS) to a volume twice the size of the pellet. Two microliters of Fc block was added to each sample and incubated for 5 min before adding Bl-Eng2 tetramer at a concentration of 5–25 nM. The tetramer stain was done for 1 h at room temperature in the dark. Samples were washed and kept on ice for the remainder of the protocol. One hundred microliters of Miltenyi anti-PE microbeads (Miltenyi 130–048–801) was added to each sample and incubated for 30 min on ice then washed and resuspended in 3 mL. LS columns were wet and samples were filtered through 40 μm filters into columns. Columns were washed with 3 mL cold sorter buffer three times before eluting bound fractions. Bound fractions were stained with invitrogen’s LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit and surface markers. Fifty microliters of AccuCheck Counting Beads was added to each sample determine total number of tetramer+ cells.
Cytokine neutralization
On the day of challenge, before yeast was given, mice received 100 μg intravenously of cytokine neutralizing antibody or rat IgG. Antibodies were given every other day until the day of harvest. Lungs were harvested at day 7 post infection, homogenized, and plated for CFU analysis on BHI agar plates. Neutralizing antibodies were purchased from BioXcell: anti-mouse IFN-γ (XMG1.2 cat#BP0055) and anti-mouse IL-17A (17F3 cat#BP0173).
CD4 depletion
Mice were vaccinated SC with Bl-Eng2 in CFA twice 2 weeks apart. Fourteen days after the last boost, mice received antibody treatment. To systemically deplete CD4+ T cells, mice received 100 μg of anti-CD4 mAb (Clone: GK1.5 Bio X Cell BE0003–1) or rat IgG as a control intravenously. To deplete lung Trm, mice received 20 μg of the same anti-CD4 antibody by intubation. Prior to challenge and 24 h after antibody treatment, a cohort of mice (n = 3 mice/group) was harvested to assess successful depletion of CD4+ T cells. Five minutes before euthanasia mice were given 2 µg fluorochrome-labeled anti-CD45 mAb as described above. Lungs, peripheral blood, spleens, and draining lymph nodes were harvested from all mice, processed, and stained to identify CD4+CD44hi lymphocytes. To assess resistance, mice were challenged with 2 × 104 B. dermatitidis yeast 24 h after antibody treatment. Six days post infection, lungs were harvested, collagenase digested, plated for lung CFU and stained with Bl-Eng2 tetramer and surface antibodies.
FTY720 inhibitor
Mice were vaccinated SC with Bl-Eng2 in GCPs or CFA + Bl-Eng2. MSA in GCP and CFA alone served as unvaccinated controls. Fourteen days following the last boost mice were injected i.p. with 25 μg FTY720 inhibitor in PBS daily to prevent T cell egress from SLOs. Two days after the first FTY720 injection, mice were challenged with 2 × 104 B. dermatitidis yeast. At day 6 post infection, lung CFU and the number of CD4+CD44hi lymphocytes in the peripheral blood were determined.
Adoptive transfer
C57BL6 wild-type mice were vaccinated SC with Bl-Eng2 in CFA twice 2 weeks apart. Two weeks after the boost, migratory T cells from the donor spleen and Trm from the lungs were harvested. CD4+ T cells from the spleens of naive mice were harvested as a negative control. Lungs were processed as described above and lymphocytes enriched by a percoll gradient. Leukocytes from individual lungs were pooled and transferred i.v. into TCR- α knockout mice on a 1–1 organ basis (ten lungs harvested → transferred into ten recipients). Spleens were mashed through 40 μm filters and red blood cells lysed. Following lysis, splenocytes were enriched for CD4+ T cells using BD iMagTM anti-mouse CD4 magnetic particles (Clone: GK1.5, BD 551539). CD4+ enriched splenocytes were then pooled, resuspended in PBS, and transferred i.v. into TCR- α knockout mice on a 1–1 organ basis (ten spleens harvested → transferred into ten recipients). Recipients received ~8 × 106 lung cells or ~20 × 106 CD4+ T cells from the spleen.
Statistics
All statistics for FACs samples were calculated in Prism (GraphPad). An unpaired two-tailed t test was used to calculate significance between vaccine groups or between untreated vs. treated groups in either absolute number or percentage of cells (Figs. 2–7). A one way ANOVA with Tukey’s test for multiple comparisons was used to calculate significance within each organ of untreated and treated mice in the CD4 depletion experiment (SFig. 8A). A Kruskal–Wallis test with Dunn’s multiple comparisons was used for analysis of the CFU data. For comparisons of cell counts, data were log transformed before t-tests were conducted. A p value of <0.05 was considered statistically significant.
References
Schenkel, J. M. & Masopust, D. Tissue-resident memory T cells. Immunity 41, 886–897 (2014).
Mueller, S. N. & Mackay, L. K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016).
Gasper, D. J. et al. Effective respiratory CD8 T-Cell immunity to influenza virus induced by intranasal carbomer-lecithin-adjuvanted non-replicating vaccines. PLoS Pathog. 12, e1006064 (2016).
Gebhardt, T. et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009).
Jiang, X. et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 483, 227–231 (2012).
Ariotti, S. et al. T cell memory. Skin-resident memory CD8(+) T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105 (2014).
Mackay, L. K. et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl Acad. Sci. USA 109, 7037–7042 (2012).
Sakai, S. et al. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J. Immunol. 192, 2965–2969 (2014).
Sallin, M. A. et al. Th1 differentiation drives the accumulation of intravascular, non-protective CD4 T cells during tuberculosis. Cell Rep. 18, 3091–3104 (2017).
Schenkel, J. M. et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014).
Shin, H. & Iwasaki, A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature 491, 463–467 (2012).
Teijaro, J. R. et al. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514 (2011).
Wu, T. et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95, 215–224 (2014).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Hofmann, M. & Pircher, H. E-cadherin promotes accumulation of a unique memory CD8 T-cell population in murine salivary glands. Proc. Natl Acad. Sci. USA 108, 16741–16746 (2011).
Masopust, D. et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med 207, 553–564 (2010).
Holmgren, J. & Czerkinsky, C. Mucosal immunity and vaccines. Nat. Med 11, S45–S53 (2005).
Kim, S. H. & Jang, Y. S. The development of mucosal vaccines for both mucosal and systemic immune induction and the roles played by adjuvants. Clin. Exp. Vaccin. Res 6, 15–21 (2017).
Lycke, N. Recent progress in mucosal vaccine development: potential and limitations. Nat. Rev. Immunol. 12, 592–605 (2012).
Wang, H. et al. C-type lectin receptors differentially induce th17 cells and vaccine immunity to the endemic mycosis of north america. J. Immunol. 192, 1107–1119 (2014).
Wang, H. et al. Ligation of Dectin-2 with a novel microbial ligand promotes adjuvant activity for vaccination. PLoS Pathog. 13, e1006568 (2017).
Wüthrich, M., Deepe, G. S. Jr. & Klein, B. Adaptive immunity to fungi. Annu Rev. Immunol. 30, 115–148 (2012).
Nelson, R. W. et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity 42, 95–107 (2015).
Wüthrich, M. et al. Calnexin induces expansion of antigen-specific CD4(+) T cells that confer immunity to fungal ascomycetes via conserved epitopes. Cell Host Microbe 17, 452–465 (2015).
Nanjappa, S. G., Heninger, E., Wüthrich, M., Gasper, D. J. & Klein, B. S. Tc17 cells mediate vaccine immunity against lethal fungal pneumonia in immune deficient hosts lacking CD4+ T cells. PLoS Pathog. 8, e1002771 (2012).
Wüthrich, M., Filutowicz, H. I. & Klein, B. S. Mutation of the WI-1 gene yields an attenuated Blastomyces dermatitidis strain that induces host resistance. J. Clin. Investig. 106, 1381–1389 (2000).
Wuthrich, M. et al. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J. Clin. Investig. 121, 554–568 (2011).
Shin, H. & Iwasaki, A. Tissue-resident memory T cells. Immunol. Rev. 255, 165–181 (2013).
Thome, J. J. et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828 (2014).
Wong, M. T. et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 45, 442–456 (2016).
Sakai, S. et al. CD4 T cell-derived IFN-gamma plays a minimal role in control of pulmonary Mycobacterium tuberculosis infection and must be actively repressed by PD-1 to prevent lethal disease. PLoS Pathog. 12, e1005667 (2016).
Wilk, M. M. et al. Lung CD4 tissue-resident memory T cells mediate adaptive immunity induced by previous infection of mice with Bordetella pertussis. J. Immunol. 199, 233–243 (2017).
Wijeyesinghe, S. & Masopust, D. Resident memory T cells are a Notch above the rest. Nat. Immunol. 17, 1337–1338 (2016).
Allen, A. C. et al. Sustained protective immunity against Bordetella pertussis nasal colonization by intranasal immunization with a vaccine-adjuvant combination that induces IL-17-secreting TRM cells. Mucosal Immunol. 11, 1763–1776 (2018).
Hondowicz, B. D., Kim, K. S., Ruterbusch, M. J., Keitany, G. J. & Pepper, M. IL-2 is required for the generation of viral-specific CD4(+) Th1 tissue-resident memory cells and B cells are essential for maintenance in the lung. Eur. J. Immunol. 48, 80–86 (2018).
Oja, A. E. et al. Trigger-happy resident memory CD4(+) T cells inhabit the human lungs. Mucosal Immunol. 11, 654–667 (2018).
Beura, L. K. et al. CD4(+) resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J. Exp. Med. 216, 1214–1229 (2019).
Smith, N. M. et al. Regionally compartmentalized resident memory T cells mediate naturally acquired protection against pneumococcal pneumonia. Mucosal Immunol. 11, 220–235 (2018).
Turner, D. L. et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7, 501–510 (2014).
Turner, D. L. et al. Biased generation and in situ activation of lung tissue-resident memory CD4 T cells in the pathogenesis of allergic asthma. J. Immunol. 200, 1561–1569 (2018).
Beura, L. K. et al. T cells in nonlymphoid tissues give rise to lymph-node-resident memory T cells. Immunity 48, 327–338 e325 (2018).
Casey, K. A. et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188, 4866–4875 (2012).
Cepek, K. L. et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature 372, 190–193 (1994).
Hoft, S. G. et al. The rate of CD4 T cell entry into the lungs during Mycobacterium tuberculosis infection is determined by partial and opposing effects of multiple chemokine receptors. Infect Immun 87, (2019). https://www.ncbi.nlm.nih.gov/pubmed/30962399
Wüthrich, M., Filutowicz, H. I., Warner, T. & Klein, B. S. Requisite elements in vaccine immunity to Blastomyces dermatitidis: plasticity uncovers vaccine potential in immune-deficient hosts. J. Immunol. 169, 6969–6976 (2002).
Wüthrich, M. et al. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J. Clin. Investig. 121, 554–568 (2011).
Galdino, N. A. L. et al. Depletion of regulatory T cells in ongoing paracoccidioidomycosis rescues protective Th1/Th17 immunity and prevents fatal disease outcome. Sci. Rep. 8, 16544 (2018).
de Araujo, E. F. et al. The IDO-AhR axis controls Th17/Treg immunity in a pulmonary model of fungal infection. Front Immunol. 8, 880 (2017).
Caminschi, I., Lahoud, M. H., Pizzolla, A. & Wakim, L. M. Zymosan by-passes the requirement for pulmonary antigen encounter in lung tissue-resident memory CD8(+) T cell development. Mucosal Immunol. 12, 403–412 (2019).
Dillon, S. et al. Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J. Clin. Investig. 116, 916–928 (2006).
Hung, C. Y. et al. Glucan-chitin particles enhance Th17 response and improve protective efficacy of a multivalent antigen (rCpa1) against pulmonary Coccidioides posadasii infection. Infect. Immun. 86, e00070–18 (2018). pii.
Acknowledgements
This work was supported by National Institute of Health Grant AI093553 (to MW) AI035681 (to BK) and AI040996 (to BK and MW). All flow data were collected at the University of Wisconsin Carbone Center Flow Lab on the BD LSR Fortessa with the NIH Shared Instrumentation Grant 1S100OD018202–01. We thank Drs Marc Jenkins from the Center for Immunology, Department of Microbiology and Immunology at the University of Minnesota for his assistance in generating the tetramer, Suresh Marulasiddappa from the Department of Pathobiological Sciences, School of Veterinary Medicine, University of Wisconsin-Madison for helpful discussions and Nydiaris Hernandez-Santos from the Department of Pediatrics at the University of Wisconsin for expert technical assistance.
Author information
Authors and Affiliations
Contributions
Generation of data (HED, LDSD, EMK, SF, DLW, TD, GK, AA, GRO): analysis of data and statistical analysis (HED, LDSD, EMK), paper preparation (HED, MW, BSK); specialized equipment and reagent generation (TD, AA, GRO), Study design (MW and BSK), concept and financial support (MW and BSK).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dobson, H.E., Dias, L.D.S., Kohn, E.M. et al. Antigen discovery unveils resident memory and migratory cell roles in antifungal resistance. Mucosal Immunol 13, 518–529 (2020). https://doi.org/10.1038/s41385-019-0244-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-019-0244-3