Abstract
Interactions between innate antiviral factors at mucosal surfaces and HIV-1 virions contribute to the natural inefficiency of HIV-1 transmission and are a platform to inform the development of vaccine and nonvaccine strategies to block mucosal HIV-1 transmission. Tenascin-C (TNC) is a large, hexameric extracellular matrix glycoprotein identified in breast milk and genital fluids that broadly neutralizes HIV-1 via interaction with the HIV-1 Envelope (Env) variable 3 (V3) loop. In this report, we characterize the specific determinants of the interaction between TNC and the HIV-1 Env. We observed that TNC binding and neutralization of HIV-1 is dependent on the TNC fibrinogen-like globe (fbg) and fibronectin-type III (fn) domains, oligomerization, and its newly-mapped glycan structure. Moreover, we observed that TNC-mediated neutralization is also dependent on Env V3 residues 321/322 and 326/327, which surround the IGDIR motif of the V3 loop, as well the N332 glycan, which is critical to the broadly neutralizing activity of glycan-dependent V3-specific antibodies such as PGT128. Our results demonstrate a striking parallel between innate and adaptive immune mechanisms of broad HIV neutralization and provide further insight into the host protein-virus interactions responsible for the natural inefficiency of mucosal HIV-1 transmission.
Similar content being viewed by others
Introduction
With the challenge of developing a safe and highly effective HIV-1 vaccine, reduction of HIV transmission rates may rely on a combination of vaccine and nonvaccine prevention methods. HIV-1 is mucosally transmitted by sexual contact and via mother-to-child transmission (MTCT). The probability of HIV-1 sexual transmission, as assessed in a cohort of heterosexual, antiretroviral-naïve (ARV) HIV-1 discordant couples, was found to be less than 1% per sexual act.1 Furthermore, in the absence of ARV prophylaxis, <10% of breastfed infants contract infection from their HIV-1 positive mothers despite daily, chronic exposure to HIV-1 in breast milk for up to two years.2 This poor transmission efficiency of HIV-1 suggests that antiviral factors in both genital fluids and breast milk contribute to natural protection against mucosal infection in the majority of HIV exposed individuals. Characterization of these innate mucosal antiviral factors may inform the development of safe and effective HIV prophylaxis and immunization strategies to further reduce the likelihood of mucosal transmission.
Breast milk, saliva, and other mucosal fluids from uninfected individuals demonstrate inherent HIV-1 inhibitory activity, suggesting the presence of innate antiviral factors.3,4,5 Furthermore, human breast milk has been shown to abrogate oral HIV-1 transmission in BLT (bone marrow, liver, thymus) humanized mice.6 We previously identified Tenascin-C (TNC) as an endogenous broad-spectrum HIV-1 neutralizing protein present in breast milk and genital fluids.7,8 Several other innate proteins in mucosal fluids, including secretory leukocyte protease inhibitor (SLPI),3,9,10 lactoferrin,11,12,13,14 mucins,3,15 and salivary agglutinin (SAG)16,17,18,19 have also demonstrated HIV-1 inhibitory activity. Conversely, semen-derived enhancer of virus infection (SEVI) has been identified as an innate enhancer of HIV infection that promotes virion attachment to target cells.20 Importantly, despite the seemingly broad HIV-neutralizing activity of identified natural antiviral inhibitors, the molecular interactions of these proteins with HIV virions and envelope proteins are largely ill-defined.
TNC is a large hexameric extracellular matrix glycoprotein with established involvement in fetal neurodevelopment, wound healing, and cancer.21 Each monomer contains an assembly domain, a region of epidermal growth factor-like repeats (EGF-L), a region of fibronectin type III-like domains (fn), and a fibrinogen-like globe (fbg).22 TNC exists in a variety of isoforms due to both alternative splicing and post-translational modifications.23 While fn 1–8 are always present in human TNC, domains fn A–D can be alternatively spliced, enabling up to 511 theoretical isoforms.23 Human TNC contains up to 26 potential sites for N-glycosylation and up to 34 potential sites for O-glycosylation, though this number varies between isomers.24 Experiments with glycoside hydrolase enzymes have demonstrated that glycans contribute significantly to the mass of TNC.24 However, it has not been previously determined how many of the possible TNC glycosylation sites are active. Moreover, the functional significance of TNC glycosylations remains relatively undefined.24 TNC was isolated from the HIV-neutralizing fraction of antibody-depleted breast milk and found to mediate HIV-1 virion capture, block virus-epithelial cell binding, and broadly neutralize HIV-1 by binding to the HIV-1 Env variable 3 (V3) loop in a charge-dependent manner,7 demonstrating a similar HIV Env-binding epitope to that of SAG gp340.16 The function of many broadly neutralizing antibodies isolated from HIV-infected humans are dependent on interaction with HIV Env glycosylations,25,26,27 including the canonical V3 loop-directed bNab PGT128.28 Moreover, the repositioning of Env N-linked glycosylation sites facilitates virus escape of neutralization from some bnAbs.29 While glycosylation of antibodies is critical for various antibody effector functions,30 the role of glycosylation of innate antiviral factors in HIV inhibition has not been explored.
In this study, we sought to define the specific determinants of the interaction between TNC and the HIV-1 Env V3 loop. We identified critical amino acid and N-linked glycan residues in the V3 loop that are required for TNC binding and neutralization of HIV, as well as specific domains of TNC that contribute to Env binding and HIV neutralization. Additionally, we identified specific patterns of glycosylation on TNC and evaluated the role of glycans on TNC-Env interactions. Mapping the precise nature of the interaction between TNC and HIV Env will further our understanding of natural non-adaptive antiviral activity against HIV-1, which can be harnessed in nonvaccine prevention strategies that circumvent the difficult task of eliciting antibody-mediated broad neutralizing activity by vaccination.
Results
TNC HIV Env-binding domains
We first identified the HIV Env-binding domains of TNC by screening a panel of monomeric TNC domains by ELISA for binding to HIV-1 Env proteins. Notably, TNC domains fn 1–8, fn A–D, fn 6–8, and fbg (Fig. 1a) all demonstrated weak binding capacity to the HIV-1 clade B Tier 1A chronic virus MN.3 gp120 Env (Fig. 1b) and to the MN.3 variable loop 3 (V3) gp70 scaffold protein (MN.V3 gp70, Fig. 1c). We did not observe substantial differences in the binding strength of truncated monomeric TNC constructs to MN.3 gp120 Env. However, the fn 6–8 domain appeared to have a lower binding potency against MN.V3 gp70 when compared to the other TNC domains. To map oligomeric TNC-HIV Env interactions, four recombinant oligomeric TNC proteins were employed: one including all fn domains (TNC-L), one excluding the fn A–D domains (TNC-S), and truncated versions of TNC-S with exclusion of either fbg (TNC-SΔfbg) or both fn and fbg (TNC-SΔfn-fbg, Fig. 1a). We observed that TNC-SΔfbg and TNC-SΔfn-fbg did not exhibit binding to MN.3 gp120 (Fig. 1d) and demonstrated reduced binding to the V3 scaffold protein (Fig. 1e), indicating a role for both fbg and the fn domains in multivalent HIV-Env binding. The mammalian cell-expressed TNC domain constructs FNall and FNall+fbg (Fig. 1a) were also tested for binding against MN.3 gp120 (Supplementary Fig. S1e) and MN.V3 gp70 (Supplementary Figure S1f). While both constructs bound to MN.V3 gp70 at levels above the albumin control, we were only able to detect weak binding for FNall against MN.3 gp120.
TNC fn and fbg domains contain HIV Env and variable loop 3 (V3) binding domains. a Schematic of TNC full length and truncated recombinant proteins. TNC-L, TNC-S, TNC-Sffbg, TNC-SΔfn-fbg, fnALL+fbg, and fnALL were expressed in mammalian cells. fn1–8. fn A–D, fn 6–8, and fbg were expressed in bacteria. TNC fbg, fn1–8, fn 6–8, and fn A–D are capable of binding both (b) gp120 and (c) conformational V3 scaffold protein. TNC-S binding to both (d) gp120 and e) conformational V3 scaffold protein is abrogated by deletion of both fbg (TNC-SΔfbg) and fbg and the fn domains (TNC-SΔfn-fbg). Either human or bovine albumin was used as a negative control binding protein in each assay. Data is displayed as the median and range of assay replicates
To further investigate the TNC domains responsible for HIV Env interaction, binding of the oligomerized TNC proteins and monomeric TNC domains to both MN.3 gp120 and MN.V3 gp70 were measured by surface plasmon resonance (SPR). Immobilized TNC domains fbg, fn 1–8, fn A–D, and fn 6–8 all demonstrated binding to MN.3 gp120 Env and MN.V3 gp70 (Supplementary Fig. S1a, S1c). We also observed a reduction in the binding response from both the TNC-SΔfbg and TNC-SΔfn-fbg proteins relative to TNC-L and TNC-S when MN.3 gp120 was flowed over immobilized TNC proteins (Supplementary Fig. S1b). TNC-S demonstrated a higher magnitude of binding to MN.V3 gp70 than TNC-L by both ELISA and SPR, suggesting that the presence or absence of the fnA-D may impact TNC conformation and in turn impact the TNC-V3 binding affinity. In contrast to the ELISA assay, we were unable to detect a difference in binding magnitude between TNC-L and TNC-SΔfbg via SPR, which may be the result of technical distinctions between these assays (Supplementary Fig. s1d).
Variable glycosylation profiles of recombinant and purified human TNC
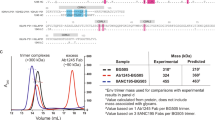
We next sought to characterize the glycosylation profile of TNC and to identify the contribution of TNC glycosylation to its HIV neutralizing activity. The recombinant human TNC-L isoform used in this study contains 23 theoretical N-linked glycosylation sites. Of these, 18 sites are located in the fn domains and one site is in the fbg domain (Supplementary Fig. S2). While it is known that human TNC is heavily glycosylated, the relative occupancy of individual TNC glycosylation sites has not previously been defined.24
To this end, we mapped the glycosylation profiles for four TNC proteins via a glycopeptide mass mapping approach, including human TNC isolated from breast milk (hTNC-L Milk) or a glioma cell line (hTNC-L Glioma) and recombinant TNC-L produced in either 293T HEK cells (rTNC-L 293T) or in 293T Lenti-X cells (rTNC-L LX). We previously observed that rTNC-L 293T did not have detectable neutralizing activity against the clade C tier 2 virus DU.156, whereas rTNC-L LX had measurable neutralizing activity similar to that of TNC purified from human milk.7 This distinction in neutralizing function suggested that post-translational modifications, such as glycosylation, may be critical to HIV-TNC interactions.
Our analysis of the high mannose and processed glycans (Fig. 2a) as well as sialylated residues (Fig. 2b) revealed considerable diversity between TNC proteins, indicating that the cell line of origin for each TNC protein has a substantial impact on its glycosylation profile. Hierarchical clustering based on profiles of high mannose glycans (Fig. 2c), processed glycans (Fig. 2d), and sialylated residues with complete assay coverage (Fig. 2e) demonstrated that the two human isolated TNC proteins clustered together and had distinct glycosylation patterns from the two recombinant TNC proteins. Notably, we did not observe features in the glycosylation profile of rTNC-L 293T that distinguished this protein from the other three neutralizing TNC proteins, indicating that glycosylation of TNC proteins is not sufficient to explain the lack of detectable neutralization activity of rTNC-L 293T.7
Glycosylation profiles of TNC and contribution of glycosylation to HIV neutralization. a High mannose and processed glycan profiles for TNC produced recombinantly in 293T HEK cells or Lenti-X 293T HEK cells, and TNC enriched from a glioma cell line or from human breast milk. b Sialylation profiles of the four TNC proteins. Heat map representations of hierarchical clustering analysis of the four TNC proteins of glycosylation site percent occupancy is displayed for c high mannose glycans (d) processed glycans and e) sialylation. f Western blot confirmation of mobility shift and glycosidase purification of rTNC following PNGase or Sialase-mediated glycan cleavage. g DU.156 neutralization profile for rTNC-L, deglycosylated, and desialated TNC-L presented as the median and range of duplicate values
The median percentage of high mannose glycans was significantly higher in the recombinant TNC proteins when compared to the purified TNC proteins, yet the two recombinant and two purified proteins did not have distinguishable high mannose glycan content when compared to each other (Supplementary Fig. S3a). Conversely, the purified TNC proteins exhibited higher median percentages of sialylated (Supplementary Fig. S3b) and processed (Supplementary Fig. S3c) residues when compared to the recombinant proteins, yet the sialylation and processed residues of two recombinant proteins and two purified proteins did not differ significantly from each other.
Enzymatic deglycosylation of rTNC-L abrogates TNC-mediated HIV neutralization
We next investigated whether bulk removal of the N-linked glycosylations of TNC altered its virus neutralization potency. We utilized PNGase F, a glycosidase that catalyzes cleavage between the innermost N-Acetylglucosamine and asparagine residues, resulting in the complete cleavage of N-linked glycosylations from TNC.31 Additionally, we utilized α2–3.6,8,9 neuraminidase A, a sialidase that cleaves terminal sialic acid residues from glycoproteins to investigate the specific contribution of sialylation to TNC-mediated virus neutralization.32 The efficiency of enzyme-mediated cleavage of TNC and subsequent purification was confirmed by western blot (Fig. 2f). In fact, both deglycosylation and desialylation of TNC abrogated neutralization activity against the clade C tier 2 HIV clinical isolate DU.156 (Fig. 2g), indicating that the presence of N-linked glycosylations on TNC may be necessary for its virus neutralization functionality, either for the structural integrity of TNC or for direct interaction. Specifically, these findings suggest that the sialic acid residues on N-linked glycosylations are necessary components for TNC virus neutralization.
TNC-mediated HIV neutralization depends on oligomerization and the fn and fbg domains
Based on our observation that deletion of TNC domains fbg and fn-fbg disrupted binding to MN.3 gp120 and MN.3 gp70 V3, we analyzed the neutralization capacity of these TNC constructs against the MN.3 virus (Fig. 3a). While TNC-L exhibited neutralization against MN.3, both TNC-SΔfbg and TNC-SΔfn-fbg failed to demonstrate neutralization activity against this virus. Furthermore, monomeric TNC domains fbg, fnALL, and fnALL + fbg all failed to neutralize HIV MN.3 (Fig. 1a, Fig. 3a), indicating that the Env-binding domains of TNC are incapable of MN.3 neutralization when displaced from the context of the intact protein.
TNC-mediated HIV neutralization is dependent on oligomerization and clade-specific usage of the fn and fbg domains. Neutralization curves for recombinant full length TNC-L, recombinant TNC proteins with domain deletions (TNC-SΔfbg and TNC-SΔfn-fbg), and monomeric TNC domains (TNCfbg, TNCfnALL, TNCfnALL + fbg) against (a) the clade B virus MN.3 and (b) the clade C virus DU.156. The median and range of quadruplicate values is displayed for rTNC-L/MN.3. The median and range of duplicate values is displayed for all other neutralization curves. c Summary of IC50 [µg/mL] values for all tested viruses
Next, we evaluated the neutralization potency of the same panel of proteins against the clade C HIV DU.156 (Fig. 3b). As with MN.3, we observed that monomeric TNC domains fbg, fnALL, and fnALL+fbg failed to neutralize DU.156. While both TNC-SΔfbg and TNC-SΔfn-fbg were able to weakly neutralize DU.156 (Fig. 3b), the average IC50 for both TNC variants was greater than 3-fold higher than that of full length rTNC-L (TNC-SΔfbg IC50 = 148 µg/mL, TNC-SΔfn-fbg IC50 = 132 µg/mL, and rTNC-L IC50 = 37.6 µg/mL), suggesting that the fbg and fn domains partially contribute to HIV-1 DU.156 neutralizing potency (Fig. 3c). To investigate whether the difference in the neutralization potencies of TNC-SΔfn-fbg and TNC-SΔfbg was clade specific, we evaluated the neutralization potencies of these constructs on two additional clade B (SF162 and CH058) and clade C (MW965 and 1086.C) viruses. Here, we observed a similar pattern of neutralization in which TNC-SΔfn-fbg and TNC-SΔfbg were completely unable to neutralize the selected clade B viruses and capable of only partial neutralization against clade C virus (Fig. 3c). This observation suggests that TNC domains other than the fn and fbg domains may be capable of HIV neutralization at a diminished potency in a clade-specific manner.
Mapping Env amino acid residue determinants of TNC binding to the HIV-1 Env V3 loop
We previously identified that TNC binds to HIV-1 gp120 at a CD4-inducible epitope, and that this binding interaction can be blocked by the V3 loop-directed mAbs (19B and F393F) and by the chemokine coreceptor binding site mAb 17b.7 On the basis of these findings, we next sought to identify specific amino acid residues in the Env V3 loop that are required for TNC binding. To this end, we utilized a panel of linear peptides of the V3 region of the MN.3 virus to identify their differential binding capacity to TNC proteins. We began by comparing the binding capacity of a 33 amino acid linear peptide corresponding to the V3 region of the wild type MN.3 virus (residues 299–331) to a 15-mer peptide consisting of residues 307 through 321 of the wild type MN.3 sequence (Fig. 4f). Full length TNC-L bound to the full length MN.V3 peptide, but did not demonstrate any binding to the 15-mer peptide (Fig. 4a). Based on this observation, we hypothesized that amino acids in the regions excluded from the 15-mer peptide are critical for binding. Therefore, we assessed TNC binding to three mutant linear MN.V3 peptides with alanine mutations introduced at positions K321A/N322A (MN.V3 321/322), T326A/I327A (MN.V3 326/327), or K321A/N322A/T326A/I327A (MN.V3 321/322/326/327) (Fig. 4f). Full length, oligomerized TNC-L demonstrated greater binding to the MN.V3 wild type peptide than to any of the alanine mutated peptides (Fig. 4a), indicating that these amino acid changes are sufficient to partially disrupt TNC binding to the linear MN.V3. Furthermore, both TNC-SΔfbg and TNC-SΔfn-fbg demonstrated no interaction with the 15-mer peptide, yet low binding to both the wild type and alanine-mutant peptides (Fig. 4b, c, f). This finding is consistent with our observation that the absence of these domains reduces binding to both MN.3 gp120 and the conformational V3 scaffold protein. Moreover, the TNC fbg and fn 1–8 domains were capable of binding to the wild type MN.V3 peptide and also demonstrated a pattern of diminished binding to the alanine-mutant peptides and no binding interaction with the 15-mer (Fig. 4d, e, f). This V3 peptide mapping thus supports the hypothesis that the binding of these domains to the V3 loop of HIV-1 Env is at least partially dependent on the amino acids in positions 332 through 337.
Amino acid residue determinants of TNC binding to linear V3. Wild type, truncated, and alanine-mutant linear MN.V3 peptides were tested by ELISA against for binding to (a) TNC-L, (b) TNC-SΔfbg, (c) TNC-SΔfn-fbg, (d) TNCfbg, and (e) TNC fn 1–8 and detected by the anti-V3 mAb CH22. The median and range of duplicate values is displayed. f Peptide sequences and heat map of ELISA AUC against TNC proteins
Mapping the TNC neutralizing epitope on HIV Env V3
Finally, based on our observation that TNC binding to linear V3 is diminished in alanine-mutant V3 peptides, we investigated the functional significance of the observed residue-specific binding on neutralization potency. To this end, we generated Env psudeotyped virions with mutations in the V3 region that corresponded to the mutations that disrupted TNC-V3 peptide binding, including mutations at 321/322 (MN.V3 K321A/N322A) and 326/327 (MN.V3 T326A/I327A). We also utilized an MN.3 pseudovirus with the glycan mutation 332 (MN.3 N332A) to investigate the role of this adjacent viral glycan in the V3 loop, which is a common target of HIV bnAb activity, in the neutralizing functionality of TNC.29
TNC-L had reduced neutralizing potency against HIV MN.3 with alanine mutations at positions 332 (IC50 = 112.53 µg/mL) and at 326/327 (IC50 = 157.6 µg/mL) when compared to the wild type virus (IC50 = 31 µg/mL) (Fig. 5a). The IC50 values for MN.3 T326A/I327A and MN.3 N332A were more than three-fold higher for the wild type virus indicating that these amino acid changes led to partial resistance of the virus to TNC-mediated neutralization (Fig. 5b). However, MN.3 K321A/N322A was not neutralized by TNC-L, indicating the amino acid residues at this position are required for the neutralization activity of TNC against HIV MN.3. Interestingly, this mapped HIV Env neutralization epitope of TNC closely matches the neutralization epitope of the human broadly neutralizing antibody PGT128, which is dependent on the N332/334 glycan as well as residue contact in the IDGIR motif from 323–327.29 PGT128 has a reported median neutralization titer of 0.01 µg/mL against a panel of clade B viruses and 0.04 µg/mL against clade C viruses. This makes PGT128 substantially more potent than TNC, for which we report neutralization titers ranging from 20.9 to 54.7 µg/mL for clade B and C viruses in Fig. 3.25 The neutralization determinant residues of TNC are highlighted on the structure of the BG505 SOSIP (Fig. 5c).33
Identification of the TNC neutralization epitope on HIV Env V3. a Neutralization curves for rTNC-L against the MN.3 Wild Type and mutant viruses MN.3 321/322, MN.3 326/327, and MN.3 332. Median and range of quadruplicate values is displayed for rTNC-L/MN.3 WT. Median and range of duplicate values is displayed for all other neutralization curves. b Summary of IC50 [µg/mL] values for the previous panel. c The defined epitope of TNC are highlighted on the structure of the BG505 SOSIP 664 trimer
Discussion
Defining the antiviral mechanisms of innate factors with HIV-inhibiting activity such as TNC will inform vaccine and nonvaccine strategies for the reduction of mucosal postnatal HIV-1 transmission. In this study, we mapped the N-linked glycosylation profile of the HIV-neutralizing mucosal fluid protein TNC and identified the terminal sialic acid residues of the N-linked glycosylations on TNC to be key to its virus neutralizing activity. We also defined TNC-Env interactions required for HIV neutralization by identifying the fbg and fn domains as the critical binding and functional domains of TNC and identifying Env V3 loop residues 321/322, 326/327, and 332 as critical for TNC-Env binding and neutralization. Remarkably, the identified neutralization epitope of TNC closely matches the previously defined neutralization epitope of the human V3 loop-binding broadly neutralizing antibody PGT128.29
Our analysis revealed that TNC fbg and fn domains are both capable of HIV-1 gp120 V3 binding. Deletion of fbg and both the fn and fbg domains dramatically reduces binding against both the conformational MN.3 gp120 HIV Env and a linearized peptide of the V3 region, suggesting similar mechanisms of TNC-HIV binding between conformational and linear HIV proteins. Alternative splicing and post-translational modifications of TNC gives rise to a number of physiological TNC isoforms.23,24 We observed that fn domains 1–8, which exist in all isoforms of TNC, are capable of HIV-1 gp120 binding, suggesting that most human TNC isoforms are capable of HIV-1 binding. While the fn A–D domains, which are present in TNC-L, but not the TNC-S splice variant, bound to gp120 and MN.V3 gp70, TNC-L and TNC-S demonstrated similar binding potencies to gp120, demonstrating the redundant contribution of these domains to the Env binding interaction and to the total virus binding capacity of TNC. However, our observation that TNC-S binds to MN.V3 gp70 with greater affinity than TNC-L may indicate that the fn A–D domain may impact TNC conformation and the binding affinity of TNC to HIV.
Not surprisingly, glycan profiles differed substantially between isolated TNC proteins purified from human cell lines or mucosal fluids (hTNC-L Milk and hTNC-L Glioma) and the two recombinantly-produced TNC proteins (rTNC-L 293T and rTNC-L 293T LX) used in this study. Both purified TNC proteins were characterized by greater amounts of sialylation and processed glycans and lower amounts of high mannose glycans when compared to the recombinant proteins, suggesting that the recombinant TNC proteins used in this study may not completely represent physiological TNC glycosylation. Yet, as rTNC-L 293T LX neutralizes HIV-1 DU.156 at a similar potency to the purified TNC proteins, the observed differences in glycosylation patterns between the recombinant and purified TNC may have only a minor impact on TNC antiviral functionality. It is interesting that the glycosylation patterns of the non-neutralizing recombinantly-produced TNC protein (rTNC-L 293T)7 did not present clear glycan differences from the three neutralizing proteins. Therefore, the observed differences in neutralization potency are less likely to be dependent on the global glycan profile and more likely the result of glycan differences at specific residues, protein conformation, or co-precipitating proteins resulting from the different production cell lines.34 We also observed that PNGase digestion of TNC disrupted neutralization of HIV-1 DU.156, indicating that TNC glycosylation may be necessary for TNC-mediated virus neutralization. However, this data does not allow us to distinguish whether TNC glycans interact directly with HIV Env to confer neutralizing activity, or if TNC glycans are necessary for the structural conformation of the protein that confers neutralizing function.
Notably, 18 of the 23 possible N-linked glycosylation sites on rTNC-L are localized to the fn domains, which we identified as the principal Env-binding region of TNC. In purified human TNC, we found that the majority of these fn glycosylation sites were heavily sialylated. Additionally, we previously identified that TNC binding to HIV-1 Env is a highly charge-dependent interaction.7 As sialic acid is negatively charged and the chemokine coreceptor binding site of Env contains a positively charged heparin-binding domain,35 we speculate that the interaction between charged sialylated residues at one or two key sites in the fn domain of TNC with positively charged regions of the chemokine coreceptor binding site of Env is a likely mechanism of TNC-mediated virus neutralization. This hypothesis is supported by the finding that neuraminidase-mediated cleavage of TNC sialylations abolished TNC-mediated neutralization activity. However, sialic acid is a hydrophilic compound that may play a role in the structural stabilization of the protein that is independent of direct sialic acid residue virus binding.
Interestingly, while deletion of the TNC fbg or the fbg and fn domains completely abrogated neutralization activity against the clade B viruses MN.3, SF162, and CH058, TNC-SΔfbg and TNC-SΔfn-fbg were each capable of weakly neutralizing the clade C viruses DU.156, MW965, and 1086.C. This finding indicates that TNC may interact distinctly with different HIV clades to mediate virus neutralization. The possibility of multiple TNC-mediated neutralization mechanisms supports our previous findings that TNC is capable of broad, cross-clade neutralization for a variety of HIV-1 strains.7,8 We also observed that monomeric TNC proteins were incapable of virus neutralization, including the fnALL+fbg monomer, which contains all of the observed Env-binding regions. This finding suggests that TNC oligomerization may be an integral determinant of virus neutralization and that polyvalency and coordination between multiple arms of TNC may be necessary for neutralizing HIV by interacting with multiple regions of the HIV Env or by interacting with multiple Env proteins on the virion surface (Fig. 6). These findings are summarized in Table S1.
Schema of the mechanism of TNC-mediated HIV-1 neutralization. TNC blocks the interaction between the HIV-1 Env and the coreceptor CCR5 / CXCR4 via binding to the HIV-1 Env V3 loop. The fn and fbg domains of individual arms of oligomerized hexameric TNC may contact multiple Envs on the virion surface
We previously determined that TNC likely neutralizes HIV-1 by binding to the Env chemokine coreceptor binding site, as evidenced by sCD4 enhancement of TNC-L:Env binding and blocking of TNC-L:Env binding by the chemokine coreceptor binding site-directed mAb 17b.7 Several amino acids in the V3 loop are predictive of coreceptor usage for both CCR5 and CXCR4-tropic viruses and characterize the coreceptor binding site on the V3 loop.36,37,38 Importantly, coreceptor usage among CCR5 and CXCR4-tropic viruses can be predicted by the identity of the amino acid residue at position 332.37Our finding that the introduction of alanine mutations at positions 321/322 of HIV-1 MN.3 abrogates TNC-mediated neutralization and fn and fbg domain binding supports that TNC neutralizes HIV-1 by blocking the chemokine coreceptor binding site in a manner that is dependent on these critical residues. Furthermore, we observed that alanine mutations at 326/327 and 332 partially disrupted virus neutralization, indicating that TNC may make additional points of contact with these residues. Notably, the canonical V3 loop-directed broadly neutralizing antibody PGT128 utilizes several of the same residues mapped as key to TNC-mediated neutralizing activity to broadly neutralize HIV, suggesting a compelling parallel between innate and adaptive immune mechanisms of broad HIV neutralization.
In conclusion, we mapped the determinants of interaction between the innate glycoprotein TNC and the HIV Env, elucidating the strategies of virus neutralization used by a natural non-adaptive antiviral factor present in mucosal fluids including glycan interactions and V3 loop determinants similar to that of V3 loop-directed bnAbs. As vaccine elicitation of a protective antibody-mediated neutralization response remains elusive, further studies exploring innate antiviral factors such as TNC can inform novel strategies for the prevention of HIV transmission and expand our understanding of the natural non-adaptive antiviral strategies utilized by mucosal fluids to yield inefficient mucosal HIV-1 transmission.
Materials and Methods
More detailed descriptions of experimental methods provided in Supplemental Methods.
Preparation of recombinant proteins
Recombinant TNC-L was produced by transfection of either 293T cells or 293T Lenti-X cells with the pEE14-HxB.L expression vector using the jetPRIME transfection reagent (Polyplus Transfection) and purified by ammonium sulfate precipitation method.7,39 TNC-SΔfbg, TNC-SΔfn-fbg, fnALL+fbg and fnALL were produced by transfection of HEK293T cells with the pHLSec2 vector, which has a histidine tag for purification. The rest of the recombinant proteins, fn 1–8, fn A–D, fn 6–8 and fbg, were expressed in Escherichia coli BL21 (DE3) with the pET15b expression vector which also has a histidine tag. His-tagged proteins were purified with a cobalt column using standard procedures.40 In the case of fbg, the protein was insoluble and required renaturation. Total purified protein was quantified by Bradford assay (Bio-Rad).
Detection of TNC protein binding to V3 peptides by ELISA
Enzyme-linked immunosorbent assays (ELISA) were performed as previously described.8 384-well ELISA plates (Corning Life Sciences) were coated with TNC proteins (TNC-S, TNC-L, fn 1–8, fn 6–8, fn A–D, fbg, FNall, FNall+fbg) (Fig. 1a), human albumin, or bovine serum albumin at 100 µg/ml in 0.1M NaHCO3 and incubated overnight at 4 °C. Blocking was performed using Super Block (1X PBS, 4% whey protein, 15% goat serum, 0.5% Tween 20) for 1 h at RT. HIV-1 Env peptides (including MN.3 gp120, MN.3 gp70 V3, and the linear peptides MN.V3, MN.V3 15-mer, MN.V3 321/322, MN.V3 326/327, and MN.V3 321/322/326/327) were titrated using a two-fold serial dilution in Super Block starting at 500 µg/ml for the linear peptides, 1,280 µg/mL for MN.3 gp120, and 1,000 µg/ml for MN.3 gp70 V3. An HIV-1 gp120 specific monoclonal antibody (CH22 or 16H3) was used as a primary antibody while detection was done using goat anti-human or anti-mouse HRP labeled (1:5,000) as a secondary antibody. Signal was then detected with SureBlue Reserve 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (VWR) and absorbance recorded at 450 nm using a Spectramax Plus spectrophotometer (Molecular Devices).
Surface plasmon resonance
Full length and truncated TNC proteins were immobilized by amine coupling to Surface Plasmon Resonance (SPR) sensor chip CM5 between 500 and 5500 response units (RU) as described previously.7,41 HIV-1 MN.3 gp120 Env protein was passed over the chip at 50 µl/min and binding was measured with a BIAcore 4000 (GE Healthcare). Nonspecific binding was subtracted over a surface immobilized with an anti-RSV IgG mAb (Synagis).
Glycosylation profile analysis
Mass spectrometry-based glycosylation profile analysis was performed as described previously42,43 and is described in detail in the Supplemental Methods.
Enzymatic deglycosylation of TNC proteins
Deglycosylation of TNC proteins was performed using Remove-iT PNGaseF or chitin-binding domain (CBD)-α2–3,6,8,9 Neuraminidase A (New England Biolabs) as per manufacturer’s protocol. Complete deglycosylation was confirmed by Western blot using an anti-TNC monoclonal antibody (clone T2H5; Abcam). Purity of deglycosylated TNC was confirmed by a reduced Coomassie stained SDS-PAGE and Western blot with an anti-CBD mAb (New England Biolabs).
Hierarchical clustering of TNC proteins based on glycosylation site occupancy
Hierarchical clustering was performed using the R programming language (Version 3.4.1, 2017; R Foundation for Statistical Computation44) on the two recombinant and two human isolated TNC proteins. The hclust function was used for clustering and dendrogram visualization on a matrix of the percent of glycosylation site occupancy for each of the 23 potential TNC glycosylation sites. Heatmaps were constructed in GraphPad Prism (GraphPad Software).
Synthesis of MN V3 Peptides
The MN V3 complete V3 loop peptide (5′–TRPNYNKRKRIHIGPGRAFYTTKNIIGTIRQAH–3′) and MNV3 15-mer peptide (5′–KRIHIGPGRAFYTTK–3′) were obtained from the NIH AIDS Reagent Program. Mutated MNV3 peptides were synthesized by CPC Scientific, Inc, including the MNV3 321/322 Mutant (5′–TRPNYNKRKRIHIGPGRAFYTTAAIIGTIRQAH–3′), the MNV3 326/327 Mutant (5′–TRPNYNKRKRIHIGPGRAFYTTKNIIGAARQAH–3′), and the MNV3 Quad Mutant (5′–TRPNYNKRKRIHIGPGRAFYTTAAIIGAARQAH–3′).
Neutralization assay
Neutralization titers were measured by the reduction in Tat-regulated Luc reporter gene expression in a TZM-bl (NIH AIDS Reagent Program) reporter cell assay, as previously described.7 Briefly, TNC proteins were prepared in serial 3-fold dilutions and incubated with virus at approximately 50,000 relative luminescence units (RLU) in 96-well flat-bottom culture plates for one hour at 37 °C. TZM-bl reporter cells, which express Firefly Luciferase in response to infection through its control by the HIV tat promoter, were added at 104 cells per well in a 100-µl volume and incubated for 48 h at 37 °C. Following 48 h incubation at 37 °C, samples were incubated in Bright-Glo luciferase reagent (Promega) for 2 min at room temperature. The resulting cell lysate was transferred into a 96-well black solid plate and RLUs were determined using a Victor X3 Multilabel Plate Reader. The median tissue culture infectious dose (TCID50) was calculated as the protein concentration required to produce a 50% RLU reduction when compared to a virus-only positive control and a cell-only negative-control. Neutralization curves are displayed as the median and range of quadruplicate values for rTNC-L vs. MN.3 WT and as the median and range of duplicate values for all other curves.
Preparation of MN.3 V3 Mutant Viruses
Mutants of the MN.3 envelope gene were constructed using the Quick Change II Site-Directed Mutagenesis kit (Agilent Technologies) following manufacturer’s instructions. Briefly, PCR was performed by Pfu DNA polymerase using B.MN.3 envelope gene cloned in pcDNA3.1D/V5-His-TOPO vector as a template DNA and corresponding primer pair was used to introduce mutation. PCR was followed by the DpnI enzyme treatment of the PCR mix at 37 °C for 1 h. This DpnI-treated PCR mix was transformed into STBL2 supercompetent cells. Twelve clones were picked for each mutant and then checked by restriction digestion analysis. Mutations were confirmed by sequencing, using an automated DNA sequencer (ABI prism). Virus infectious titers were measured using TZM-bl reporter cells. The resulting virus from the MN.V3 321/322/326/327 plasmid was found to be not infectious. Primer sequences are as follows. MNV3 321/322/326/327 For: 5′–GCATTTTATACAACAGCAGCTATAAAAGGAGCTGCAAGACAAGCACATTG–3′; MNV3 321/322/326/327 Rev: 5′- CAATGTGCTTGTCTTGCAGCTCCTTTTATAGCTGCTGTTGTATAAAATGC–3′; MNV3 321/322 For: 5′–GCATTTTATACAACAGCAGCTATAAAAGGAACTATAAGAC–3′; MNV3 321/322 Rev: 5′–GTCTTATAGTTCCTTTTATAGCTGCTGTTGTATAAAATGC–3′; MNV3 326/327 For: 5′–CAAAAAATATAAAAGGAGCTGCAAGACAAGCACATTG–3′; MNV3 326/327 Rev: 5′-CAATGTGCTTGTCTTGCAGCTCCTTTTATATTTTTTG–3′.
Structure generation
TNC binding site structure figure was generated in the PyMol Molecular Graphics System, Version 1.8, Schrödinger, LLC45 using the structure model of ligand free BG505 SOSIP 664 (pdb 4zmj).33
Statistical analysis
The nonparametric Kruskal-Wallis test was used to compare TNC glycosylation profiles. Subsequent pairwise comparisons were performed with the Dunn’s multiple comparisons test. All statistical tests were performed with Graphpad (La Jolla, CA) Prism version 6.
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Gray, R. H. et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357, 1149–1153 (2001).
Coutsoudis, A. et al. Late postnatal transmission of HIV-1 in breast-fed children: an individual patient data meta-analysis. J. Infect. Dis. 189, 2154–2166 (2004).
Kazmi, S. H. et al. Comparison of human immunodeficiency virus type 1-specific inhibitory activities in saliva and other human mucosal fluids. Clin. Vaccine Immunol. 13, 1111–1118 (2006).
Fox, P. C., Wolff, A., Yeh, C. K., Atkinson, J. C. & Baum, B. J. Saliva inhibits HIV-1 infectivity. J. Am. Dental Assoc. 116, 635–637 (1988).
Malamud, D., Davis, C., Berthold, P., Roth, E. & Friedman, H. Human submandibular saliva aggregates HIV. AIDS Res. Hum. Retroviruses 9, 633–637 (1993).
Wahl, A. et al. Human breast milk and antiretrovirals dramatically reduce oral HIV-1 transmission in BLT humanized mice. PLoS Pathog. 8, e1002732 (2012).
Fouda, G. G. et al. Tenascin-C is an innate broad-spectrum, HIV-1-neutralizing protein in breast milk. Proc. Natl Acad. Sci. USA 110, 18220–18225 (2013).
Mansour, R. G. et al. The presence and anti-hiv-1 function of tenascin c in breast milk and genital fluids. PLoS ONE 11, e0155261 (2016).
Farquhar, C. et al. Salivary secretory leukocyte protease inhibitor is associated with reduced transmission of human immunodeficiency virus type 1 through breast milk. J. Infect. Dis. 186, 1173–1176 (2002).
Shugars, D. C., Alexander, A. L., Fu, K. & Freel, S. A. Endogenous salivary inhibitors of human immunodeficiency virus. Arch. Oral. Biol. 44, 445–453 (1999).
Newburg, D. S., Viscidi, R. P., Ruff, A. & Yolken, R. H. A human milk factor inhibits binding of human immunodeficiency virus to the CD4 receptor. Pediatr. Res. 31, 22–28 (1992).
Berkhout, B. et al. Characterization of the anti-HIV effects of native lactoferrin and other milk proteins and protein-derived peptides. Antiviral Res. 55, 341–355 (2002).
Moriuchi, M. & Moriuchi, H. A milk protein lactoferrin enhances human T cell leukemia virus type I and suppresses HIV-1 infection. J. Immunol. 166, 4231–4236 (2001).
Takayama, Y., Aoki, R., Uchida, R., Tajima, A. & Aoki-Yoshida, A. Role of CXC chemokine receptor type 4 as a lactoferrin receptor. Biochem. Cell Biol. 95, 57–63 (2017).
Mthembu, Y. et al. Purified human breast milk MUC1 and MUC4 inhibit human immunodeficiency virus. Neonatology 105, 211–217 (2014).
Wu, Z., Golub, E., Abrams, W. R. & Malamud, D. gp340 (SAG) binds to the V3 sequence of gp120 important for chemokine receptor interaction. AIDS Res. Hum. Retroviruses 20, 600–607 (2004).
Wu, Z. et al. Salivary agglutinin inhibits HIV type 1 infectivity through interaction with viral glycoprotein 120. AIDS Res. Hum. Retroviruses 19, 201–209 (2003).
Chu, Y., Li, J., Wu, X., Hua, Z. & Wu, Z. Identification of human immunodeficiency virus type 1 (HIV-1) gp120-binding sites on scavenger receptor cysteine rich 1 (SRCR1) domain of gp340. J. Biomed. Sci. 20, 44 (2013).
Patyka, M., Malamud, D., Weissman, D., Abrams, W. R. & Kurago, Z. Periluminal distribution of HIV-binding target cells and gp340 in the oral, cervical and sigmoid/rectal mucosae: a mapping study. PLoS ONE 10, e0132942 (2015).
Münch, J. et al. Semen-derived amyloid fibrils drastically enhance hiv infection. Cell 131, 1059–1071 (2007).
Orend, G. & Chiquet-Ehrismann, R. Tenascin-C induced signaling in cancer. Cancer Lett. 244, 143–163 (2006).
Midwood, K. S. Advances in tenascin-C biology. Cell. Mol. Life Sci. 68, 3175–3199 (2011).
Midwood, K. S. Tenascin-C at a glance. J. Cell. Sci. 129, 4321–4327 (2016).
Giblin, S. P. M. & Kim, S. Tenascin-C: form versus function. Cell Adh. Migr. 9, 48–82 (2015).
Walker, L. M. et al. Broad neutralization ocverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Trkola, A. et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70, 1100–1108 (1996).
Bonsignori, M. et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med. 9, eaai7514 (2017).
Pejchal, R. et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011).
Krumm, S. A. et al. Mechanisms of escape from the PGT128 family of anti-HIV broadly neutralizing antibodies. Retrovirology. 13, 8 (2016).
Raju, T. S. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr. Opin. Immunol. 20, 471–478 (2008).
Tarentino, A. L. & Plummer, T. H. Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230, 44–57 (1994).
Iwamori, M., Ohta, Y., Uchida, Y. & Tsukada, Y. Arthrobacter ureafaciens sialidase isoenzymes, L, M1 and M2, cleave fucosyl GM1. Glycoconj. J. 14, 67–73 (1997).
Kwon, Y. D. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015).
Brellier, F. et al. SMOC1 is a tenascin-C interacting protein over-expressed in brain tumors. Matrix Biol. 30, 225–233 (2011).
Crublet, E., Andrieu, J. P., Vives, R. R. & Lortat-Jacob, H. The HIV-1 envelope glycoprotein gp120 features four heparan sulfate binding domains, including the coreceptor binding site. J. Biol. Chem. 283, 15193–15200 (2008).
Cashin, K. et al. Covariance of charged amino acids at positions 322 and 440 of HIV-1 Env contributes to coreceptor specificity of subtype B viruses, and can be used to improve the performance of V3 sequence-based coreceptor usage prediction algorithms. PLoS ONE 9, e109771 (2014).
Resch, W., Hoffman, N. & Swanstrom, R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288, 51–62 (2001).
Huang, C. C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 (2005).
Aukhil, I., Joshi, P., Yan, Y. & Erickson, H. P. Cell- and heparin-binding domains of the hexabrachion arm identified by tenascin expression proteins. J. Biol. Chem. 268, 2542–2553 (1993).
Shah, R., Ohashi, T., Erickson, H. P. & Oas, T. G. Spontaneous unfolding-refolding of fibronection type iii domains assayed by thiol exchange. J. Biol. Chem. 292, 955–966 (2017).
Alam, S. M. et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178, 4424–4435 (2007).
Go, E. P. et al. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J. Proteome Res. 8, 4231–4242 (2009).
Go, E. P. et al. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes’ accessibility. J. Proteome. Res. 7, 1660–1674 (2008).
Ihaka, R. G. R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 (1996).
The PyMOL Molecular Graphics System, Version 1.8. (Schrödinger, LLC, New York, NY, 2015).
Acknowledgements
We would like to thank David Montefiori for providing the MN.3 N332A mutant virus; David Martinez, Amit Kumar, and Holly Heimsath for technical support and advice; Nathan Nicely for generation of the Env trimer structure figure; and Maria Blasi and Mary Klotman for providing us with the 293T Lenti-X cell line. Protein antigens for ELISAs were generously provided by Kevin Saunders and Barton Haynes. Production of the antigens was supported by NIH, NIAID, Division of AIDS UM1 grant AI100645 for the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID). The following reagent was obtained through the NIH AIDS Reagent Program, AIDS Program, NIAID, NIH: HIV-1 MN Complete V3 Loop Peptide, and TZM-bl cell line. This work was funded by HHS | National Institutes of Health (5R01-DE025444; R01GM103547).
Author information
Authors and Affiliations
Contributions
R.J.M. and S.R.P. designed and analyzed experiments, interpreted the data, and wrote the manuscript; L.S. designed and performed experiments and analysis and contributed to writing the manuscript; J.A.E. performed and analyzed neutralization experiments; T.O. and H.E. generated recombinant TNC proteins; FHJ, B.W., and S.M.A. performed and analyzed the SPR experiments; H.L.I. performed virus production; G.G.F. provided insight for the interpretation and discussion of results; E.P.G. and H.D. designed and analyzed the glycan mapping and provided insight for interpretation and discussion. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mangan, R.J., Stamper, L., Ohashi, T. et al. Determinants of Tenascin-C and HIV-1 envelope binding and neutralization. Mucosal Immunol 12, 1004–1012 (2019). https://doi.org/10.1038/s41385-019-0164-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-019-0164-2