Abstract
Asthma comprises heterogeneous clinical subtypes driven by diverse pathophysiological mechanisms. We characterized the modulation of the inflammatory environment with the phenotype, gene expression, and function of helper CD4 T cells among acutely exacerbated and stable asthma patients. Systemic Th2 immune deviation (IgE and Th2 cytokines) and inflammation (IL-6, CRP) were associated with increased Th17 cells during acute asthma. Th2/Th17 cell differentiation during acute asthma was regulated by the enhanced expression of transcription factors (c-MAF, IRF-4). The development of pathogenic Th2 cells during acute asthma was characterized by the secretion of inflammatory cytokines coupled with Th2 molecules and PPARγ expression. The acquisition of CD15S, CD39, CD101, and CCR4 contributed to the increased heterogeneity of Regulatory T cells during asthma. Two clusters were derived from above cytokines, CD4 T cell phenotypes, and clinical data. Cluster 1, characterized by high eosinophils, Th2 and ILC2 frequencies, and higher exacerbation rates, may represent Th2-high subtype. Cluster 2 represents a more complex subtype; it is constituted by higher neutrophils or Th17 frequencies, higher inhaled corticosteroids dose and poor asthma control. In conclusion, we characterized systematically and longitudinally Th2-high and non-Th2 asthma subtypes and the heterogeneity of CD4 T cells in stable and acute asthma.
Similar content being viewed by others
Introduction
There is an emerging consensus that asthma is a heterogeneous disease that comprises of multiple clinical subtypes.1,2,3 The pathophysiology of asthma may involve a variety of innate and adaptive cells2 including type 2 innate lymphoid cells (ILC2),4 Th2, Th17,5 regulatory T cells (Tregs)6, and follicular helper T (TFH) cells.7 Moreover, interactions between the innate and adaptive immune systems may be crucial for the initiation and propagation of the immune responses in asthma.8
Th2 cells contribute to hallmark features of asthma mainly via the production of type 2 cytokines, including IL-4, IL-5, and IL-13, which are further regulated by the specific expression of GATA-3. The recent discovery of the ability of ILC2 to secrete large quantities of type 2 cytokines allows us to infer their potential in mediating asthma pathophysiology, and recent findings in mice9 further support a role for the innate immune system in driving asthma. In humans, a higher frequency of ILC2 was detected in patients with allergic asthma as compared to healthy individuals or even patients with allergic rhinitis.10 The observation of higher frequencies and activity of ILC2 in the human lung9 and in mouse models of airway inflammation11,12 confirmed their influential role in driving lung tissue pathologies. Asthmatic individuals, who demonstrate higher blood and airway eosinophilia, bronchial hyper responsiveness, and atopy,13 have been classified as Th2-high subtype patients. Immune therapies targeting Th2 cytokines such as IL-5 (mepolizumab) are therefore effective in reducing asthma exacerbation and systemic corticosteroid dependency in Th2-high but not in non-selected asthmatic patients.14,15 Allergen-specific Th2 cells (CCR6−CCR4+CRTH2+) are highly differentiated and can be characterized by the expression of homing receptors CD49d and CD161. The combined secretion of inflammatory cytokines (TNF-α, GM-CSF, IL-9) with classical Th2 molecules could explain their pathogenic role in driving asthma progression. This allergen-driven Th2 effector program is regulated by the transcription factor PPARγ, either intrinsically or extrinsically in T cells and dendritic cells respectively.16,17
Th2-low (or non-Th2) subtype represents a more heterogeneous group of patients. Nevertheless, Th2 responses are less frequent in non-atopic donors and their symptoms reveal a greater involvement of Th17 cells such as increased IL-17A secretion,18 CD161,17 and PPARγ expression.16 Despite their indispensable role in aiding allergen clearance and maintaining mucosal barriers, observations in mice models demonstrate a role for Th17 cells in driving neutrophilic19,20 or eosinophilic19,21 airway inflammation as well as in promoting bronchial hyper-reactivity.5,19 Moreover, asthma patients were found to have elevated IL-17A levels in plasma22,23 and possess higher frequencies of IL-17+ CD4 T cells24 in the lungs. Furthermore, IL-17A levels correlated with the disease severity of neutrophilic, steroid-resistant asthma.22,23 Although the secretion of antibodies (immunoglobulin E (IgE)) is regulated by the activity of TFH, few studies have examined the role of TFH in asthma exacerbation and those that exist present conflicting results.7 Independently of asthma, the increased frequencies of Th2-like TFH cells have been linked to allergic rhinitis with or without asthma25 and the development of food allergies.26 IL-21, which is in part secreted by TFH cells, was reported to both promote27 and suppress28 the IgE response in different mouse models of allergic airway inflammation.
Th2 cell activities are finely modulated by the actions of regulatory T cells, which are recruited to the effector site29 (through the up-regulation of chemokine receptors such as CCR4 and CCR830) and limit the tissue-damage that would otherwise be caused by other inflammatory T cells. The lack of immune regulation by Tregs is associated with pathologic Th2 response,31 and the induced hyper-responsiveness of Tregs (through mucosal exposure to nonpathogenic antigens) could potently suppress Th2-mediated responses.32 Hence, a clearer understanding of the mechanisms involved could yield therapeutic opportunities for treating asthma.
Overall, observations in mice demonstrate that specific interactions between multiple T helper subsets and innate cells have critical roles in driving asthma pathogenesis and have given rise to a great heterogeneity of asthma phenotypes. However, these findings have not been validated in acute asthmatic patients. Through the immunological characterization of a clinically well-defined longitudinal cohort of stable and acute asthma young adult patients (free from other chronic complications), the present study relies on an unsupervised and high-throughput approach to comprehensively describe the dynamics and specific involvement of major adaptive CD4 T cell subsets including Tregs, Th1, classical and non-classical Th2, Th17, TFH CD4 T cells, and innate population ILC2 in different asthma settings. These immunological signatures are further linked to the expression of specific cytokines and transcription factors. Via various cutting-edge clustering algorithms, we were able to assign distinct immunological profiles and clinical outcomes to the different asthma subtypes, further corroborating the existing data from animal model and pilot clinical studies.
The unique immunological signatures of individual asthma subtypes highlight the complex pathophysiology of asthma and raise useful implications for personalized therapy.
Results
The baseline demographic and clinical profiles of study participants are shown in Table S1. by acute asthma, stable asthma, and healthy control groups. There were no significant differences in mean age, proportions of gender, and family housing conditions among the three groups (p > 0.05). Participants with acute asthma were more likely to be smokers (p < 0.001) and have lower levels of lung function (forced expiratory volume in one second (FEV1) predicted and forced vital capacity (FVC) predicted, p < 0.01). Compared to patients with stable asthma, those with acute asthma had significantly longer mean duration of asthma history of 18.9 versus 15.1 years (p < 0.001); they did not have significantly more numbers of asthma attacks, but were more frequently hospitalized in the last 12 months. Six (46.2%) patients in the acute asthma group and 20 (42.6%) patients in the stable asthma group reported taking a medium to high dose of ICS (p = 0.82). Patients with acute asthma gave lower asthma control scores (13.23 ± 5.12 vs. 18.86 ± 4.75, p < 0.001), and poorer asthma-related quality of life (3.94 ± 1.49 vs. 5.10 ± 1.35, p < 0.01).
Plasmatic cytokine signature of acute asthma patients
Firstly, we investigated if exacerbation of asthma could induce a specific Th2 immune deviation. We studied plasma samples from our cohort of acute and stable asthma and healthy donors (HD) at baseline and during longitudinal follow-up. Higher levels of IgE (p values = 0.0001 and <0.001), IL-10 (p values = 0.0057 and =0.0222), TSLP (p values = 0.0385 and =0.0064), IL-4 (p values < 0.0001 and =0.0015) and IL-13 (p values < 0.0001 and =0.0006) were detected in acute and stable asthma groups respectively compared with HD. Inflammatory molecules such as IL-6 (acute asthma vs. HD: p < 0.0001; stable stable vs. HD: p = 0.0008) and CRP (acute asthma vs. HD: p = 0.0006; stable asthma vs. HD: p = 0.0026) were also exacerbated during asthma manifestation, particularly in the acute asthma group (Fig. 1a, d). Cytokines and IgE concentration was reduced in the acute group during 1-year follow-up (Fig. 1b, e), concomitantly with the normalization of clinical parameters (lung function, medication usage, asthma severity). The stable asthma group had generally minimal changes of clinical data at 6-month and 12-month, although the changes were more apparent than in the healthy controls (Fig. 1c, f). Elevated IL-2 during acute asthma (p < 0.001) might reflect indirectly the dynamic activation of T cells.
The increase of peripheral Th2-associated cytokine levels in asthma patients. a Radar representation of cytokines concentration (pg/ml) detected in the plasma from acute (n = 20) or stable asthma (n = 46) patients and the healthy controls (n = 69) at baseline; b Longitudinal follow-up of plasmatic cytokines in the acute asthma group at 6-month and 12-month; c Longitudinal follow-up of plasmatic cytokines in the stable asthma group at 6-month and 12-month. d Increased Inflammatory Th2 response at baseline and during follow-up in acute (e) or stable patients (f). The cytokine concentrations were measured by Luminex (IgE, IL-4, IL-13, IL-10, IL-25, IL-6) or ELISA (TSLP, CRP). Statistical analysis was performed in comparison to control group (a) or to baseline (b, c). Significant differences were indicated by asterisks; * p < 0.05, ** p < 0.01, *** p < 0.001
Cellular signature of acute asthma patients
To gain insight into the modulation of the cellular composition during asthma exacerbation, we enumerated longitudinally the frequencies of innate and adaptive immune cells. Although there was a trend for differences in multiple innate leukocyte sub-populations, only eosinophil frequency was significantly higher in stable asthma patients than in HD (p = 0.026). In mixed model for repeated measures, acute and stable asthma was associated with increased frequencies of iNKT (identified as TCR Vα24Jα18+ CD45,+ p = 0.001), eosinophils (identified as CCR3+CD66+CD45,+ p < 0.001), and neutrophils (identified as CCR3−CD66+CD45,+ p = 0.038). Stable chronic asthma was also linked to elevated frequency of basophils (p = 0.035) (Table S2).
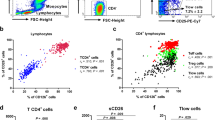
Innate γ/δ T cells were decreased during asthma (Fig. 2a). Adaptive immune T lymphocytes were also deeply modulated during Asthma. The frequency of CD4 T cells was increased whereas CD8 T cells subsets were reduced (Fig. 2b), and consequently a higher CD4/CD8 ratio (Fig. 2c) than HD. This proportion was preserved during the 1-year follow-up of acute patients (Fig. 2b, d).
Stable increase of CD4 T cells during acute and stable asthma. The frequencies of α/β and γ/δ T cells (a) or CD4 and CD8 T cells (b) were measured by multi-parameter flow cytometry in freshly isolated PBMCs at baseline (n = 119) in patients with acute or stable asthma or healthy controls. Baseline (c) and longitudinal follow-up (d) of CD4/CD8 ratio measured in patients with acute asthma. e Signatures of acute and stable asthma at baseline and after 1 year of follow-up were represented by log median changes
The differentiation stage of T cells was also impacted during asthma. Central memory T cells (CM, CD62L+CD95+) are the main T cell lymphocytes with long term memory properties and are essential during recall response against pathogens. However, it is unknown yet if these cells are or are not detrimental during asthma. As shown in Table S3, we found that acute asthma was associated with an enrichment of CM CD4 but decrease of CM CD8 T cells. The most differentiated CD8 T cells were defined as CD45RA+CD27− terminal effector (TE) T cells and were decreased in all asthma patients at the initiation of the study. In the re-assessment at 6-month and 1-year, memory T cells remained elevated during asthma in comparison to HD. We could wonder if the persistence of allergens may explain the enhancement of the memory response.
We summarized the main clinical and immunological parameters during acute and stable asthma in comparison to HD at the recruitment time and after 1-year of follow-up in Fig. 2e. Inflammatory Th2 response, decreased lung function (FEV1 predicted, FVC predicted) and a neutrophil-mediated disease characterized the acute manifestation of asthma. Peripheral increase of eosinophils was apparent only during stable asthma. After one year of follow-up most of the significant variables modulated during acute asthma (except IL-4 and CRP) were normalized in comparison to stable asthma.
Modulation of helper CD4 T cells during asthma
Th2 plasmatic signature coupled with the increased frequency of CM CD4 T cells supported the hypothesis of massive proliferation and differentiation of memory Th2 during asthma. It was thus investigated whether the source of Th2 cytokines were adaptive Th2 cells or innate ILC2. The composition of CD4 T cells during asthma was generated using t-SNE33,34 which utilizes unbiased clustering to visualize high-dimensional data. The identification of regulatory and helper T cells enabled us to visualize classical and non-classical subsets during acute and stable asthma (Fig. 3a). The clusters specific to Th1 (C8, C10), Th2 (C15), Th17 (C5, C14), TFH (C13), and Tregs (C16) were modulated during asthma (Fig. 3b). The intensity of some markers also changed during asthma. CD127 expression in Th2 cells might explain their proliferation (Fig. 3c).
Homeostasis of helper CD4 and regulatory T cells during acute asthma. a CD4 T cell composition during Asthma. t-Distributed Stochastic Neighbor Embedding (t-SNE) was generated by Cytofkit among concatenated representative patients with acute or stable asthma and healthy controls. The expression of each marker was automatically scaled. b Differentiation of CD4 T cells during acute and stable asthma. c Characterization of CD4 T cells during asthma. Representative heat map of the fluorescence intensity in clusters from healthy controls, acute, and stable asthma patients. d Identification of Helper CD4 T cells during asthma. Th1, Th17, and Th1-Th17 subsets were identified as CCR6−CXCR3+, CCR6+CXCR3−, and CCR6+CXCR3+ respectively. e Depletion of Th1 and increased frequency of peripheral Th17 populations during acute asthma. Comparison of the frequencies of Helper CD4 T cells among the healthy control group, the acute asthma group, and the stable asthma group (n = 72) was performed by flow cytometry. f Inversion of Th1/Th2 and Th1/Th17 ratio during acute asthma. g Increased CCR4 expression on Th2 and Th17 CD4 T cells during acute asthma. Statistical analysis of CCR4 MFI in Helper CD4 T cells was performed among the healthy control group, the acute asthma group, and the stable asthma group (n = 72)
The identification of Th2 was performed by the combination of surface markers, such as CCR4 and CRTH2, and intra-cellular expression of specific Th2 transcription factor GATA-3 in healthy controls and acute or stable asthma patients (from left to right) (Fig. 3d). Surprisingly, the Th2 frequency was not altered in peripheral blood of asthma patients (Fig. 3e). However, they presented lower Th1/Th2 ratio than HD (p < 0.01 and p = 0.06 for stable and acute asthma vs. HD respectively; Fig. 3f) and a specific increase of CCR4 expression (p < 0.05) during acute asthma (and enhancement of GATA-3 expression in Th2 cells from stable patients (data not shown)) (Fig. 3g). We wondered if ILC2 could contribute to Th2 deviation during asthma despite its low frequency in peripheral blood. Patients with stable asthma had lower ILC2 frequency but elevated CCR4 expression than other groups. GATA-3 was decreased in ILC2 of acute asthma patients (p < 0.05; Fig. S1). ILC2 (CD127+CD161+CD25+CRTH2+GATA-3+) represented the main ILC population (CD127+CD161+CD25+T-betlow) in our cohort but we also detected with unbiased clustering the existence of others ILC subsets characterized by CCR6 and CXCR3 expression.
We sought to determine which environmental factors could contribute to or limit manifestation of acute asthma. The high prevalence of CMV infection in our cohort and the positive correlation of Th1 frequency with CMV-associated response prompted us to examine if this persistent infection could influence the nature of immune response during asthma. Th2 frequency negatively correlated with CMV IgG level in stable asthma (rho = −0.713, p = 0.009) and HD (rho = −0.607, p = 0.028) (Table S4). The correlation between CMV IgG and frequencies of Th1 and Th2 cells were not performed in acute asthma patients due to the small number of patients in the group who had both data available. Dedicated studies are needed to determine whether Th1 immune deviation associated with CMV response may limit pathology due to inflammatory Th2 cells during asthma.
We dissected further helper T cell composition during asthma through a differential combination of chemokine receptors in order to define Th1 (CCR6−CXCR3+), Th17 (CCR6+CXCR3−), and Th1-Th17 (CCR6+CXCR3+) (Fig. 3d). Increased Th17 frequencies were observed during acute and stable asthma compared with HD (p < 0.01 and p < 0.05 respectively; Fig. 3e). Consequently, Th1/Th17 ratio was significantly decreased in acute and stable asthma (p < 0.001; Fig. 3f). Acute asthma was also characterized by increased frequency of Th1-Th17 cells (p < 0.01 and p < 0.001 in comparison to control and stable asthma respectively). As observed for Th2, CCR4 expression was increased as a surface marker of Th17 during acute asthma (Fig. 3g).
The last major CD4 helper T cells studied were TFH cells. No difference in absolute TFH frequency (identified as CXCR5+ CD4 T cells) among the groups was detected. TFH cells were also diversified in term of chemokine receptor expression (and associated function). Acute asthma patients presented higher frequencies of TFH1-17 (CCR6+CXCR3+) and TFH17 (CCR6+CXCR3−) than other groups (Fig. S2).
Gene expression and functions of helper CD4 T cells during asthma
We hypothesized that asthma could induce quantitative and qualitative modulation of T cell functions. Th1, Th2, Th17, and TFH cells were sorted (Fig. S4) and polyclonally stimulated to compare their activities during acute and stable asthma (Fig. S5). The levels of Th1 (IL-2, TNF-α, and IFN-γ), Th2 (IL-5, IL-9, IL-10, IL-21, IL-17E/IL-25, IL-31, and IL-33), and Th17 associated cytokines (IL-17A, IL-17F, GM-CSF, and IL-22) are shown in Fig. 4a. The global identity of each subset of helper CD4 cells was preserved during asthma. The normalized heat map was utilized to visualize the fold change of median cytokine concentration. If the activity of Th1 was reduced during asthma, the secretion of cytokines by Th2 cells was increased for IL-5, IL-9, IL-10, IL-13, as well as IL-21 and IL-22. Some cytokines were differentially released by Th2 cells during acute asthma (IFN-γ, TNF-α, IL-17F). The function of Th17 cells were also enhanced during asthma for classically Th17associated molecules such as IL-17A and IL-22 but also for non-classical molecules including IL-5 and IL-13. The increased frequency of TFH17 during acute asthma was characterized by increased secretion of IL-17F and IL-22.
Activation of non-classical transcriptional pathways and related cytokines during acute allergic asthma. a Heat map (blue to red) of normalized fold change (vs. the healthy control group) of median cytokine concentration secreted by helper T cells after TCR stimulation with a-CD3/CD28 microbeads. Sorted Th1, Th2, Th17 and TFH cell populations were stimulated and incubated (37 °C; 5% CO2) for 48 h. Supernatants were collected and examined for expression of cytokines in Th1, Th2, Th17, and TFH cells using Luminex assay. b Heat map of normalized fold change of transcriptional factors of helper CD4 T cells during asthma. Poly-clonally stimulated sorted Th1, Th2, Th17, and TFH cells were lysed and analyzed for their transcription factor expression by RT-PCR. Fold change of each transcription factor expression in acute or stable asthma was calculated versus its corresponding expression in CD4 T cell subsets from control donors
The differentiation and acquisition of effector functions in helper CD4 T cells are regulated by concerted actions and expression of transcription factor (TF). We sought to determine if changes of cytokine signature during acute asthma were coupled with the alteration of TF expression (Fig. 4b and Fig. S6). We normalized the gene expression with the mRNA level of each TF in control donors. As expected, the master key TBX21 (gene coding for T-bet), GATA-3, and RoRγt were specifically detected in Th1, Th2, and Th17 respectively (Fig. S6). However, these expressions were not exclusive and we observed a more complex pattern of TF expression during asthma. Bcl-6, Foxo3 and IRF4 were down-regulated in Th2 whereas c-MAF was slightly increased during acute asthma. IRF4 was particularly interesting. It appears critical for the acquisition of effector T cell functions and its loss compromised Th2 differentiation due to direct regulation of IL-4, GATA-3 and Gfi1.35 We could wonder if this pattern of expression could be due to inflammation or whether it could reflect heterogeneity of Th2 cells. The gene expression of other helper CD4 T cells was also deeply altered during asthma. Th1 displayed up-regulation of unrelated Th1 TF such as RoRγt and Bcl6. Th17 was stable except for the decreased expression of IRF4, Foxo3, and PRDM1 (Blimp-1). At this stage, we could only speculate that the immune deviation observed during asthma (IL-22, IL-5, etc.) may result from a reprogramming of Th17 cells and might lead to a reduced pathogenicity of Th17.36 Finally, the TFH signature was also perturbed during acute asthma. The down-regulation of TFH-specific TF Bcl-6 was coupled with up-regulation of Th17 genes (RoRγT, c-MAF). These results suggest a development of TFH17 profile during acute asthma compatible with the expression of CCR6 by TFH cells.
The Th2 polarization of immune cells during asthma is thus characterized by a qualitative change of gene expression, phenotype, and cytokine secretion. At this stage, we could wonder if the heterogeneity within Th2 subsets could explain the differential role of pathogenic and non-pathogenic Th2 in the pathology of asthma.18,37,38,39
Induction of pathogenic Th2 cell subsets during acute asthma
The recent identification of allergen-specific Th2 cells in atopic individuals prompted us to investigate in the presence of these pathogenic Th2 cells during acute asthma.17 In order to compensate for the absence of tetramers in our study, we used the described phenotype to enumerate inflammatory Th2 cells. We excluded any potential contaminations by Th17 or Th9 CD4 T cells by gating out CCR6-expressing CD4 T cells.
The allergic T cell signature was confirmed in our cohort and included the coexpression of CRTH2, CD161, and CD49d (Fig. 5a). These Th2 cells were highly differentiated (CD45RO+CD27−) but did not express the senescence marker CD57 (data not shown). As shown in Fig. 5b, a significant increase of CD161+CRTH2+ Th2 was detected in acute and stable asthma (p = 0.0012 and p = 0.0019 respectively). The frequency of classical CRTH2 Th2 was also enhanced but only in acute asthma (p = 0.0076). The main Th2 subset (CCR4+CRTH2−CD161−) remained unchanged in our patients. We did not observe any striking differences in term of the pathogenic Th2 phenotype during acute asthma. The global signature of these inflammatory Th2 cells was thus preserved (Fig. 5c). We sought to determine if the functions of these cells could explain their deleterious role during asthma. First, we sorted and polyclonaly stimulated classical CCR4+CRTH2− and CCR4+CRTH2+ CD4 T cells, CCR4+CD161+CRTH2+ and Th17 CD4 T cells from three healthy donors. Due to the relative low frequency of inflammatory Th2 cells, a large volume of blood was required. This prevented us to perform this experiment in allergic donors or in more healthy donors. Pathogenic Th2 cells secreted limited amount of IFN-γ and IL-17A. As expected, the secretory profile of pathogenic Th2 cells was enriched in Th2associated cytokines such as IL-4 (p = 0.0283), IL-5 (p = 0.0908), IL-13 (p = 0.0332) in comparison to CCR4+CRTH2- T cells. Inflammatory signature was defined by the presence of TNF-α and GM-CSF in the absence of IL-10. The ability of CRTH2 cells to secrete IL-2 was enhanced in comparison to classical Th2 cells. Finally, a strong trend of an increased production of IL-9 in inflammatory Th2 was observed but was not statistically significant (most likely due to the number of donors) (Fig. 5d).The regulation of pathogenic Th2 cells has been recently described as PPARγ and IL-4/IL-33 dependent16,17 in human studies and animal models. We were able to demonstrate the expression of two isoforms of this TF in activated CD161+CRTH2+ CD4 T cells (Fig. 5e). The modulation of PPARγ expression and pathogenic Th2 cell activity may potentially be used as a new therapeutical target during inflammatory asthma.
Induction of pathogenic Th2 cells during acute asthma. a Heterogeneity of Th2 cells during asthma. Representative density plots of Th2 (CCR6−CCR4+) subsets identified by the differential expression of CD161 and CRTH2 in healthy donors (n = 25), acute (n = 8) and stable (n = 8) asthma patients. Th2 populations from acute asthma patients were overlaid on the histograms in order to compare the relative expression of CD49d, CD27, and GATA-3. b Increase of non-classical Th2 cells during asthma. Frequencies of Th2 subsets were assessed in frozen PBMCs from healthy donors and acute or stable asthma patients. c Phenotype of pathogenic Th2 cells during Asthma. Median fluorescent intensity of surface and intra-cellular markers was compared between gated Th2 subsets in all donors. d Increased effector function of non-classical Th2 cells during asthma. Th2 subsets and Th17 cells were sorted and polyclonally stimulated with PMA/ Ionomycin. Supernatants were collected and analyzed by luminex to detect cytokine secretion. e Expression of the transcription factor PPARγ in pathogenic Th2 cells. Sorted T cells were lysed after stimulation. The levels of mRNA PPARγ isoforms were analyzed by RT-PCR in classical or pathogenic Th2 and Th17 CD4 cells
Increased diversity of regulatory T cells during asthma
The proliferation and cytokine secretion of T cells are controlled by Tregs to limit tissue damage during prolonged immune responses. Tregs defects might also explain the exacerbated Th2 immune response against the allergen. Tregs were strictly defined as CD127lowCD25highFoxp3+ CD4 T cells (Fig. S3A). Tregs frequency and Foxp3 expression were preserved during acute and stable asthma. Tregs/Th17 ratio from acute asthma was borderline but statistically insignificant (p = 0.066). Next, we investigated if asthma was associated with specific phenotype of Tregs. The heterogeneity of regulatory T cells was identified during asthma using UMAP unbiased analysis on concatenated samples from acute and stable asthma patients and healthy controls (Fig. 6a). These non-classical Tregs included differential expression of molecules such as CCR4, CCR10, CXCR3, CD15S, CD39, and CD101 (Fig. 6b, c). Several clusters were defined by phenographs and few were particularly enriched for CD15S, CD101 and CD39 expression during asthma (Fig. 6c and Fig. S3). In order to avoid any contaminations by non-suppressive effectors cells (potentially expressing Foxp3 after activation), Tregs were stringently defined as CD127lowFoxp3+ Helios+ CD4 T cells. An increased frequency of Tregs co-expressing CD15S and CD39 (p = 0.0172 and p = 0.0056 in acute and stable asthma respectively) or CD101 (p = 0.004 and p < 0.001 in acute and stable asthma respectively) was observed during asthma (Fig. 6d). This accumulation was even more pronounced during stable asthma than in acute astma for CD15S+CD101+ Tregs (p = 0.0124). Moreover, Tregs need to migrate to the same location as effector helper T cells to specifically mediate their inhibitory roles,29,40 and therefore have to share some characteristics such as TF and chemokine receptor expression profile. If the profile of Tregs homing and migration differed during stable asthma as assessed by the decreased expression of CCR4 (Fig. S3), we noticed that CD39+ Tregs could present an increased expression of CCR4 during acute asthma (p = 0.035). Interstingly, CCR10 was also up-regulated on CD39+ Tregs during acute (p = 0.022) and stable asthma (p = 0.044). CD39,41,42 CD15S43, and CD10144,45 have been described as key molecules to identify highly supppressive Tregs. CD39+ Tregs were able to control allergic airway inflammation in an animal model46 but were reduced during allergic asthma.47 We wondered if the up-regulation of these markers during asthma was associated with an enhancement of Tregs activity. We performed co-culture experiments on isolated Tregs (defined as CD127lowCD25high) and effector CD4 T cells (defined as CD127+CD25−) from frozen PBMCs of patients with stable asthma or healthy controls (Fig. 6e). The number of cells and the availability of samples limited our approach to study only a ¼ ratio of Tregs/Teff. The CFSE dilution of effector CD4 T cells was decreased in presence of regulatory T cells in asthma patients and healthy controls. The inhibition of T cell proliferation was evaluated by the frequency of live non-dividing CD4 T cells (Fig. 6f). The frequency of non-proliferating CD4 T cells was increased in presence of Tregs from healthy controls (p = 0.0003) and stable asthma patients (p = 0.0023). However, no significant differences were observed regarding the suppressive functions of Tregs during asthma.
Heterogeneity of regulatory T cells during asthma. a UMAP repartition of Tregs from acute and stable asthma patients and healthy controls. Concatenated Tregs (CD127lowFoxP3+Helios+) from acute (n = 10) and stable asthma patients (n = 13) and healthy controls (n = 9) were visualized by UMAP. Similar number of events from the different groups was represented. Density plots represented the relative frequency of Tregs. b Phenotype of Tregs during asthma. Concatenated samples from acute and stable asthma patients and healthy controls were analyzed for their relative expression of surface and intracellular markers. c UMAP repartition of Tregs markers and of clusters during asthma. Normalized intensity of each marker was represented. Phenograph determined automatically individual clusters, which were represented by specific colors. d Frequency of non-classical Tregs during Asthma. The subsets of Tregs defined as CD127lowFoxP3+Helios+ CD4 T cells were quantified in frozen PBMCs from acute (n = 10) and stable asthma patients (n = 13) and healthy controls (n = 9). Frequencies were analyzed by Mann–Whitney U test. e Suppressive functions of Tregs during asthma. Representative histogram plots of CD4 T cell co-culture. CFSE-stained effector CD4 T cells (CD127+CD25−) were stimulated with a-CD3/CD28 beads alone or in presence of Tregs (1/4 ratio) during 4 days. f Preserved functions of Tregs during asthma. Paired t-tests were used to compare the frequencies of non-proliferating effector CD4 T cells. Test significances were indicated by asterisks; *p < 0.05, **p < 0.01, ***p < 0.001
Modelization of network analysis, specific signatures of acute and stable asthma, and identification of asthma subtypes
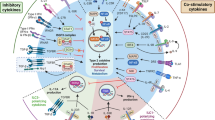
We modeled the relationship between TFs and cytokines during asthma with String Software (Fig. 7a). High confidence interaction score helped us to represent and understand the action types (activation, inhibition) between the different molecules. K-means clusters defined four main clusters from the protein network in an unbiased approach. Inflammatory molecules were associated with Th17 family members whilst Th1 and Th2 were closely related in an exclusive manner. Indeed, TFs (TBX21 and GATA-3) and cytokines (IFN-γ versus IL-4) inhibited each other reciprocally. In order to summarize our data, we tried to establish a signature of acute and stable asthma that integrates cytokines detected in the plasma and secreted by helper CD4 T cells, composition and phenotype of immune cells, and asthma-related clinical parameters (Fig. 7b). The increased heterogeneity of Th2 cells may explain the enhancement of their functions during asthma. The close monitoring of inflammatory molecules, pathogenic Th2 and Th17 cells may be informative in assessing therapy responsiveness. Together, these data imply that inflammation and immune differences between acute and stable asthma are essential to the clinical manifestations of the disease.
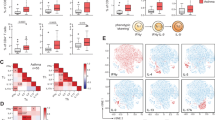
Pathway analysis and differential signature of asthma. a Network analysis of pathways associated with asthma. Analysis was performed using STRING, which is a biological database and web resource of known and predicted protein–protein interactions. All nodes in this network are colored nodes representing query proteins and first shell of interactors. The colors of the nodes represent clusters determined by K-means clustering. Edges represent protein-protein functional associations. String representation summarized the documented interactions between transcription factor and cytokines. An annotation score ≥ 0.5 indicates a high confidence interaction. b Differential signature of acute and stable asthma at baseline was represented by volcano plot. Data relative to clinical parameters, peripheral cytokines, phenotypes, and functions of helper T cells were integrated on this representation. The x-axis and y-axis show log2 (fold change) of the values for the acute asthma group versus the stable asthma group and the corresponding –log10 (p value) respectively. Dots in blue indicate higher value in the stable asthma group and in red indicate higher value in the acute asthma group. Labels were attached to significant parameters falling above the line of –log10 (0.05). Labels in red, yellow, green, blue, and purple are for chemokine receptors, peripheral cytokines, cytokines secreted by cell subsets, clinical parameters, and phenotyping respectively. c Asthma subtypes identified by cluster analysis using all parameters of immune phenotyping, cytokines, and clinical data. Data were standardized before analysis. Clusters were plotted in the first principal components by principal component analysis (PCA). Each dot represents one asthma patient. Dots in blue are those in cluster 1 and in red are those in cluster 2. PC principal component. d Heat map of cluster centers of each standardized parameter in cluster analysis. Blue indicates relatively low values and red indicates relatively high values. MFI mean fluorescence intensity, TE terminal effector, CM central memory, ICS inhaled corticosteroids (3 = medium to high dose ( ≥ 250 μg/day), 2 low dose ( < 250 μg/day), 1 = no use), ACQ asthma control questionnaire, FEV1 Pred forced expiratory volume in 1 second predicted, FVC Pred forced vital capacity predicted
Finally, cluster analysis was performed on asthma patients using all parameters of blood composition, phenotyping of immune cells, systemic cytokines, and clinical data stated as above in order to identify potential asthma subtypes. The optimal number of clusters was determined by the Mclust package of R software based on Bayesian information criterion (BIC). As shown in Fig. S7, the largest BIC appeared in Mclust VVI (diagonal, varying volume and shape) model with 2 clusters. K-means clustering was then performed with the optimal number (i.e., 2) of clusters. Among the 48 patients without any missing data for the included parameters, there were 29 patients (60.4%) in cluster 1, and 19 patients (39.6%) in cluster 2. The distributions of the two clusters were depicted in Fig. 7c. The standardized score and absolute values of cluster centers of each parameter in cluster analysis was shown in Fig. 7D. and Table S5 respectively. Cluster 1 was characterized by elevated eosinophil count, Th2 and ILC2 frequencies, and asthma exacerbation rates. It may thus represent Th2-high asthma subtype. IgE did not discriminate finely patients in this cohort. The level of IgE was increased in both clusters (cluster 1 center: 2598.66 pg/ml, cluster 2 center: 4309.76 pg/ml), and especially in cluster 2. Cluster 2 showed high neutrophil count, Th17 frequency and inverted Th1/Th17 ratio, and inhaled corticosteroids dose, poor asthma control (ACQ score) and lung function (FEV1 Pred (%) and FVC Pred (%)) and concomitant detection of high Th1 (IL-2), Th2 (IL-4, IL-13), and inflammatory cytokines (IL-6). Cluster 2 can be considered as a non-Th2 asthma subtype which represents the complexity and heterogeneity of asthma.
Discussion
Corroborating the recent findings of the diverse contribution of multiple adaptive and innate immune cells to the heterogeneity of asthma, the present study derived two clusters that correspond either to a Th2-high or a non-Th2 asthma subtype. These findings are based on the analysis of donor blood composition - i.e the detailed characterization of immune cells, systemic cytokines, and relevant clinical parameters. This study also shows that in addition to classical Th2 cells, asthma is also associated with an increased frequency of non-classical regulatory and helper T cells.17,48,49 These results have important implications towards the formulation of personalized therapies against asthma.
The immune-modulation of innate populations may represent one of the earliest biological events that initiate the secondary phase of the immune response. In our cohort, we identified a role for innate cells in driving the pathophysiology of asthma. The infiltration of eosinophils and neutrophils50 has been recognized as a hallmark feature of asthma, and is respectively characteristic of a Th2 high and non-Th2 asthma subtypes in this clinical study. In comparing the two asthma subtypes, the frequency of ILC2 was elevated in patients from the Th2-high subtype. Although the hierarchical importance of the contribution of Th2 and ILC2 in the pathology of asthma remains unknown, we could speculate that the release of Th2 cytokines by ILC2 contributes to a biochemical environment that sensitizes the adaptive immune response towards allergen reactivity.10,51,52,53 We did not have access in this study to lung biopsy or broncho-alveolar lavage fluid to evaluate the tissular response and compare the local and peripheral immune responses in different asthma subtypes. Whether the homing and migration potential to tissue (lung) of Th2 and ILC2 are similar and related to CCR4 and GATA-3 modulation will require further investigation.
The nature and extent of cytokine secretion by T cells not only determines the rate of elimination of allergens/pathogens, but also the amount of bystander activation during inflammation. In a type 2-infection model, the convergence of chromatin landscapes (i.e. regulomes) between ILC2 and Th2 cells was previously described54; it may therefore be worthwhile to determine whether the epigenetic signatures of these cellular subsets are similarly linked in Th2-high asthma, since this knowledge is useful in directing therapeutic interventions. Nevertheless, increased IL-4, IL-13, and IL-2 levels were observed even in non-Th2 asthma subtype, indicating the complexity and heterogeneity of non-Th2 asthma. Although greater attention has been given to the function of IL2 in driving T-cell proliferation and thereby contributing to sustained pathogenic Th2 activities, IL-2 is also shown to provide migrational signals to allergen-specific T cells during asthma.55 The specific role of ILC2 and Th2 cells in driving the level of each Th2 cytokine therefore warrants further evaluation and corroboration in other cohort studies – so that their contribution can be carefully weighed to determine the respective dominance in each asthma subtype so as to achieve a more targeted therapeutic approach.
The present study found that the increased frequency of Th17 population was coupled with inverted Th1/Th17 ratio in the cluster of non-Th2 asthma subtype, suggesting that Th17 cells are also key players in the heterogeneity of the pathophysiology of human asthma. In mouse studies, antigen airway sensitization primes not only Th2 cellular responses, but also strong Th17 cell responses that promote airway neutrophilia and acute airway hyper-responsiveness.56,57 Since the latter is chiefly orchestrated by the expression of the master TF RoRγt,58 it was not surprising that asthma patients could be stratified into three groups (Th2high, Th17high and Th2/17low 59) based on the genetic signature of their endo-bronchial tissue. Here, we corroborate earlier findings of higher neutrophils and Th17 frequencies in non-Th2 asthma. Furthermore, Th2 and Th17 inflammatory pathways are reciprocally regulated in this study; and Th17-related cytokines such as IL-17A and IL-22 were highly expressed in non-Th2 asthma. These findings are consistent with those from previous studies that reported increased RoRγt and RoRC activity in the PBMCs of asthmatic patients.60,61 In the different asthma subtypes, the balance between RoRγt (and amplified by Foxo362) and GATA-3 expression by CD4 T cells may therefore be important in driving either the Th17- and Th2- centric neutrophilia and eosinophilia respectively.
As shown in the present study of acute and stable asthma, Th2 cells are capable of orchestrating different outcomes in asthma pathology due to their predisposition of the intrinsic and extrinsic environment. The dominance of the TF PPARγ is characteristic of pathogenic Th2 cells and supports effector function acquisition in Th2 cells (IL-5) but not Th2 differentiation (IL-4).16 Nevertheless, the tissue-specific prevalence of Th2 cytokines such as IL-4 and IL-33 may act to perpetuate allergic Th2 conditioning.63 Together with others,64 we observed that the up-regulation of c-MAF during acute asthma could be a hallmark signature of pathological Th2 cells that is triggered by allergen challenge. Next, the increased expression of multi-drug resistance type 1 membrane transporter ABCB1/MDR1 by Th2 cells during acute asthma warrants future investigation, since it can be speculated that the coupling of ABCB1 and CD161 expression has similar roles for Th2 and Th17 cells in the pathology of asthma.65
Although a better understanding of the biology of Th2 cells during asthma is in itself an important endeavor, we must be further capable of identifying and monitoring dysfunctional Th2 cells. In this study, we validated the tracking of CRTH2, CD161, CD49d, and CD27 expression in Th2 cells as a robust method of identifying allergen-responsive Th2 cells.17 Although pathogenic Th2 cells are rare, they can be enriched and therefore studied for their allergen specificity. Nevertheless, we need also further explore whether pathologic Th2 cells can be neutralized in vivo, as this could open new therapeutic opportunities for asthma patients. Regulatory T cells that express CD15S,43 CD39 and CD10144 have been shown to effectively participate in both Th246 and Th17 regulation42; increasing the frequencies of these Tregs in asthma patients may therefore present an effective treatment opportunity. The diagnostic value of monitoring CD39 expression is reinforced in studies which show that CD39 polymorphism69 and differential expression47 contribute to different risk profiles in allergic asthma and allergic rhinitis.
Overall, the present study suggests the complexity in the Th2 response, since it is heterogeneous and not restricted to the contribution of Th2 cells. Although participants were excluded from this study if he/ she had taken oral steroids within 2 weeks before the onset of this study, this study share the common features of clinical studies and could not fully exclude the potential confounding of steroids intake. The identification of mechanisms that drive the differentiation and inflammatory functions of pathogenic helper CD4 T cells may facilitate the classification of asthma subtypes and subsequent personalized treatment regimens.2 Given the diversity of T cells that are involved in the pathology of asthma, it would be naïve to expect that targeting Th2 cells alone could mitigate pathologies in asthma; it is likely that clinical interventions need consider the crosstalk between the various immune cell types involved to be effective in controlling asthma pathogenesis.
Materials and methods
Study design
This study recruited 139 young adult donors aged 21–35 years, including 50 patients with stable asthma (no asthma attack for > 3 months), 20 patients with acute asthma exacerbation, and 69 age- ( ±2 years) and gender-matched healthy controls. All donors were free from any other chronic disease. Patients with asthma had not taken oral steroids for at least 2 weeks before study onset. The participants’ demographic, asthma clinical and immunological profiles were assessed at each visit. Spirometric tests were administered and 20 milliliter venous blood samples were collected from donors into the BD Vacutainer® CPT™ Cell Preparation tube with Sodium Citrate at baseline, 6−month, and 1-year respectively. All donors signed written informed consent for the study, which was approved by the National Healthcare Group Domain Specific Review Board of Singapore. The detailed information of study design and participants can be found in the online Supplementary Materials.
Luminex and ELISA tests
Plasmatic levels of cytokines including IL-4, IL-13, IL-10, IL-25, IL-6, IL-2, and IL-12p70 were tested using Luminex kits from Millipore. The concentration of IgE and transforming growth factor beta 1 (TGF-β1) was measured by Millipore Human Single Plex. The levels of C-reactive protein (CRP), Thymic Stromal Lymphopoietin (TSLP) and IL-33 were determined using ELISA kits from Millipore, R&D Systems and ENZO respectively. Plates were run according to the manufacturers’ instructions.
Immune cell phenotyping
Whole blood (100 μl) was lysed using red blood cells (RBC) lysis buffer (eBioscience) and stained with the following antibodies (panel 1) as markers of leukocytes: CCR3, CD64, CD66b, CD45, V alpha 24 J alpha 18 TCR and CD14.
Fresh peripheral blood mononuclear cells (PBMCs, 2 million) were isolated and stained for stages of T cell differentiation using the following markers: CD3, CD4, CD8, CD27, CD45RA, CD62L, CD95, CD31, and TCR γ/δ (panel 2).
Unstained PBMCs were cryopreserved in 90% fetal bovine serum (FBS) containing 10% dimethyl sulfoxide (DMSO). On the day of surface and ICS, 2 million cells were thawed rapidly and washed extensively with PBS containing 10% FBS. Sample recovery was >75% without loss of specific immune populations. The viability of cells was >95% as tested by trypan blue exclusion. T cells and ILC2s were labeled using the following markers: CD3, CD4, CD8, CD25, CD127, CCR4, CRTH2, CCR6, CXCR3, CXCR5, CTLA-4, FOXP3, and GATA-3 (panel 3).
We used FACS Symphony to perform high-dimensional immunophenotyping and identify classical and pathogenic Th2 cells from 2 million PBMCs with the following antibodies: CD3, CD4, CD14, CD25, CD127, CCR4, CRTH2, CCR6, CXCR3, CXCR5, CD57, CD38, CD94, CD49d, CD56, TCR-Vα7.2, CD27, CD45RO, CD161, FOXP3, T-bet, Helios, GATA-3 (panel 4).
We used FACS Symphony to identify classical and non classical Tregs cells from 2 million PBMCs with the following antibodies: CD3, CD4, CD14, CD15S, CD25, CD39, CD101, CD127, CCR4, CCR6, CCR10, CXCR3, CD94, CTLA-4, FoxP3, Helios (panel 5).
The dyes and detailed information of antibodies in each panel are summarized in Table S6. All preparations included a Live/Dead marker to exclude dead cells and false positive staining. Cells were suspended in PBS + 5% FBS in a concentration of 10 million cells/ml and transferred to a U-bottom 96-well plate to reach desired cell density (2 million cells/well). For surface marker staining, the wells were added antibody mix and incubated for 30 min at 4 °C with the plate sealed with aluminum foil. Next, fixation and permeabilization were performed. Two-hundred microlitre pixel buffer (fixation/permeabilization concentrate (4×) diluted with fixation/permeabilization diluent) was added to each well and incubated for 30 min at 4 °C with the plate sealed with aluminum foil. After centrifuge and discarding supernatant, the wells were added 200 µl/well permeabilization buffer (10×, diluted with water). Finally, intracellular staining (ICS) was done by adding the antibodies to the wells and incubating for 30 min at 4 °C with the plate sealed with aluminum foil. The samples were washed, transferred to polystyrene tubes, centrifuged, and re-suspended into 200 µl PBS followed by acquisition at BD LSRFortessa™ X-20. Multicolor flow cytometry data were analyzed using FlowJo (Treestar) and FACSDiva (BD Biosciences).
Cell sorting
Thirty million PBMCs were thawed, filtered, and stained with surface markers CD3, CD4, CRTH2, CCR6, CXCR5, CXCR3, and Live/Dead (panel 6) to sort Th1, Th2, Th17, and TFH cell populations. Fifteen million PBMCs were thawed, filtered, and stained with surface markers CD3, CD4, CRTH2, CCR6, CCR4, CD161, and Live/Dead (panel 7) to sort Th2 subsets and Th17 cell. Stained cells were washed, centrifuged, and suspended in PBS + 5% FBS to obtain a concentration of 10 million cells/ml. Compensation controls were prepared including unstained cells, positive beads (binding antibody), and negative beads (not binding antibody). Cells were sorted with PBS at 3000 events/min on BD FACSAria™ Cell Sorter. Samples were collected into sterile polypropylene tubes prefilled with 500 µl of PMRI-1640 + 20% FBS.
Cell stimulation and cytokine expression
Sorted Th1, Th2, Th17, and TFH cell populations were obtained from pooled PBMCs and stimulated with CD3/CD28 human T-activator (Thermo Fisher Scientific). Cells were incubated (37 °C; 5% CO2; Tractor Supply Company) for 48 h. Supernatants were collected and examined for expression of cytokines IL-5, IL-17E/IL-25, IL-31, IL-33, IL-17A, IL-17F, GM-CSF, IL-2, IL-9, IL-10, IL-21, IL-22, TNF-α, and IFN-γ in Th1, Th2, Th17, and TFH cells using Millipore Luminex assay (HTH17MAG-14k-10).
Sorted Th2 subsets and Th17 cell were stimulated with PMA/Ionomycin. Cells were incubated overnight at 37 °C; 5% CO2 (Tractor Supply Company). Supernatants were collected and examined for expression of cytokines IL-4, IL-5, IL-13, IL-17A, GM-CSF, IL-2, IL-9, IL-10, TNF-α, and IFN-γ in Th2 and Th17 cells using Millipore Luminex assay (HTH17MAG-14k-10). All stained antibodies were from BD Biosciences, BioLegend, and Invitrogen.
Gene expression of transcription factors
Polyclonally-stimulated Th1, Th2, Th17, and TFH sorted T cells were lysed and analyzed for their TF expression by reverse transcription polymerase chain reaction (RT-PCR). RNA extraction was performed using an RNeasy Plus Micro kit (Qiagen) according to manufacturer’s instructions. Briefly, samples were first lysed and homogenized. The lysate was passed through a gDNA Eliminator spin column followed by the addition of ethanol to the flow-through, and the sample was afterwards applied to an RNeasy MinElute spin column. Only RNA binded to the membrane and contaminants were eliminated. mRNA was reverse transcribed into cDNA using the SuperScript First Strand kit (Invitrogen) according to manufacturer’s instructions. The first-strand cDNA synthesis reaction was catalyzed by SuperScript® II Reverse Transcriptase (RT). cDNA was analyzed by real-time PCR with the KAPA SYBR qPCR Master Mix kit (KAPA Biosystems) and the following primers from Qiagen: TBX21, PRDM1, RORγt, c-MAF, ABCB1, bcl6, Foxo3, IRF4, GATA-3, and BAG6, and customized primers for PPARG-1 and PPARG-2.16
CMV IgG titers
The level of CMV IgG was measured by ELISA (Genesis Diagnostics). Plates were read at 450 nm, and the concentration of CMV IgG was calculated according to OD values.
Statistical analysis
Data analysis was performed using GraphPad Prism version 6, PASW/SPSS 18.0, and R software. One-way analysis of variance (ANOVA) and Kruskal–Wallis test were performed for data with normal and non-normal distribution respectively for the comparison among patients with stable or acute asthma and the healthy control group at baseline. Mixed model and Friedman test were used to investigate the time effect within the group when applicable. Heat map of normalized fold change versus the healthy controls of median cytokine concentration in T helper subsets and volcano plots were generated using R. Composition of CD4 T cells was analyzed using t-distributed stochastic neighbor embedding (t-SNE)33,34 and automatic clustering by Matlab to visualize high-dimensional data. Composition of Tregs was analyzed using Uniform Manifold Approximation and Projection (UMAP) (https://doi.org/10.1101/298430) 69 and automatic clustering by phenograph to visualize high-dimensional data. Network analysis of pathways was performed using STRING66,67 (https://string-db.org/), a biological database and web resource of known and predicted protein–protein interactions. Post-hoc analysis was performed with Bonferroni adjustment for one-way ANOVA and Dunn adjustment for Kruskal-Wallis test and Friedman test. Asthma subtypes were identified by k-means cluster analysis using parameters of immune phenotyping, cytokines, and clinical data. Parameterized Gaussian finite mixture model was used to identify optimal number of clusters according to Bayesian information criterion (BIC). Clusters were plotted in the first principal components by principal component analysis (PCA). Statistical significance was set at p < 0.05.
References
Kuo, C. S. et al. T-helper cell type 2 (Th2) and non-Th2 molecular phenotypes of asthma using sputum transcriptomics in U-BIOPRED. Eur. Respir. J. 49, pii: 1602135 (2017).
Fajt, M. L. & Wenzel, S. E. Asthma phenotypes and the use of biologic medications in asthma and allergic disease: the next steps toward personalized care. J. Allergy Clin. Immunol. 135, 299–310 (2015). quiz 311.
Kuo, C. S. et al. A transcriptome-driven analysis of epithelial brushings and bronchial biopsies to define asthma phenotypes in U-BIOPRED. Am. J. Respir. Crit. Care Med. 195, 443–455 (2017).
Spits, H. & Di Santo, J. P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Kudo, M. et al. IL-17A produced by alphabeta T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat. Med. 18, 547–554 (2012).
Faustino, L. et al. Regulatory T cells accumulate in the lung allergic inflammation and efficiently suppress T-cell proliferation but not Th2 cytokine production. Clin. Dev. Immunol. 2012, 721817 (2012).
Varricchi, G. et al. T follicular helper (Tfh) cells in normal immune responses and in allergic disorders. Allergy 71, 1086–1094 (2016).
KleinJan, A. Airway inflammation in asthma: key players beyond the Th2 pathway. Curr. Opin. Pulm. Med. 22, 46–52 (2016).
Halim, T. Y., Krauss, R. H., Sun, A. C. & Takei, F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity 36, 451–463 (2012).
Bartemes, K. R., Kephart, G. M., Fox, S. J. & Kita, H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J. Allergy Clin. Immunol. 134, 671–678 e674 (2014).
Doherty, T. A. et al. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J. Allergy Clin. Immunol. 132, 205–213 (2013).
Kim, H. Y. et al. Innate lymphoid cells responding to IL-33 mediate airway hyperreactivity independently of adaptive immunity. J. Allergy Clin. Immunol. 129, 216–227 (2012). e211-216.
Woodruff, P. G. et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit. Care Med. 180, 388–395 (2009).
Haldar, P. et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N. Engl. J. Med. 360, 973–984 (2009).
Pavord, I. D. et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet 380, 651–659 (2012).
Nobs, S. P. et al. PPARgamma in dendritic cells and T cells drives pathogenic type-2 effector responses in lung inflammation. J. Exp. Med. 214, 3015–3035 (2017).
Wambre, E. et al. phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci. Transl. Med. 9, pii: eaam9171 (2017).
Cosmi, L. et al. Identification of a novel subset of human circulating memory CD4( + ) T cells that produce both IL-17A and IL-4. J. Allergy Clin. Immunol. 125, 222–230 (2010). e221-224.
Manni, M. L. et al. Molecular mechanisms of airway hyperresponsiveness in a murine model of steroid-resistant airway inflammation. J. Immunol. 196, 963–977 (2016).
Fogli, L. K. et al. T cell-derived IL-17 mediates epithelial changes in the airway and drives pulmonary neutrophilia. J. Immunol. 191, 3100–3111 (2013).
Wakashin, H. et al. IL-23 and Th17 cells enhance Th2-cell-mediated eosinophilic airway inflammation in mice. Am. J. Respir. Crit. Care Med. 178, 1023–1032 (2008).
Molet, S. et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J. Allergy Clin. Immunol. 108, 430–438 (2001).
Al-Ramli, W. et al. T(H)17-associated cytokines (IL-17A and IL-17F) in severe asthma. J. Allergy Clin. Immunol. 123, 1185–1187 (2009).
Pene, J. et al. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J. Immunol. 180, 7423–7430 (2008).
Takhar, P. et al. Class switch recombination to IgE in the bronchial mucosa of atopic and nonatopic patients with asthma. J. Allergy Clin. Immunol. 119, 213–218 (2007).
Coeffier, M., Lorentz, A., Manns, M. P. & Bischoff, S. C. Epsilon germ-line and IL-4 transcripts are expressed in human intestinal mucosa and enhanced in patients with food allergy. Allergy 60, 822–827 (2005).
Coquet, J. M. et al. Interleukin-21-producing CD4( + ) T cells promote type 2 immunity to house dust mites. Immunity 43, 318–330 (2015).
Shang, X. Z. et al. IgE isotype switch and IgE production are enhanced in IL-21-deficient but not IFN-gamma-deficient mice in a Th2-biased response. Cell. Immunol. 241, 66–74 (2006).
WFt, Carson et al. RS. Accumulation of regulatory T cells in local draining lymph nodes of the lung correlates with spontaneous resolution of chronic asthma in a murine model. Int. Arch. Allergy Immunol. 145, 231–243 (2008).
Saito, K. et al. Differential regulatory function of resting and preactivated allergen-specific CD4 + CD25 + regulatory T cells in Th2-type airway inflammation. J. Immunol. 181, 6889–6897 (2008).
Shi, H. Z. & Qin, X. J. CD4( + )CD25( + ) regulatory T lymphocytes in allergy and asthma. Allergy 60, 986–995 (2005).
Keller, A. C. et al. Hierarchical suppression of asthma-like responses by mucosal tolerance. J. Allergy Clin. Immunol. 117, 283–290 (2006).
Acuff, N. V. & Linden, J. Using visualization of t-distributed stochastic neighbor embedding to identify immune cell subsets in mouse tumors. J. Immunol. 198, 4539–4546 (2017). (Baltimore, Md: 1950).
Geoffrey E. Hinton S. T. R. Stochastic Neighbor Embedding. Part of: Advances in Neural Information Processing Systems 15 (NIPS 2002), 2002.
Huber, M. & Lohoff, M. IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 44, 1886–1895 (2014).
Jain, R. et al. Interleukin-23-induced transcription factor blimp-1 promotes pathogenicity of T helper 17 cells. Immunity 44, 131–142 (2016).
Endo, Y. et al. Eomesodermin controls interleukin-5 production in memory T helper 2 cells through inhibition of activity of the transcription factor GATA3. Immunity 35, 733–745 (2011).
Islam, S. A. et al. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5 + T(H)2 cells. Nat. Immunol. 12, 167–177 (2011).
Mitson-Salazar, A. et al. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human T(H)2 cell subpopulation with enhanced function. J. Allergy Clin. Immunol. 137, 907–918.e909 (2016).
Lloyd, C. M. & Hessel, E. M. Functions of T cells in asthma: more than just TH2 cells. Nat. Rev. Immunol. 10, 838–848 (2010).
Borsellino, G. et al. Expression of ectonucleotidase CD39 by Foxp3 + Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 110, 1225–1232 (2007).
Kared, H., Fabre, T., Bedard, N., Bruneau, J. & Shoukry, N. H. Galectin-9 and IL-21 mediate cross-regulation between Th17 and Treg cells during acute hepatitis C. PLoS. Pathog. 9, e1003422 (2013).
Miyara, M. et al. Sialyl Lewis x (CD15s) identifies highly differentiated and most suppressive FOXP3high regulatory T cells in humans. Proc. Natl Acad. Sci. USA 112, 7225–7230 (2015).
Schey, R. et al. CD101 inhibits the expansion of colitogenic T cells. Mucosal Immunol. 9, 1205–1217 (2016).
Fernandez, I. et al. CD101 surface expression discriminates potency among murine FoxP3 + regulatory T cells. J. Immunol. 179, 2808–2814 (2007).
Li, P. et al. CD39 + regulatory T cells attenuate allergic airway inflammation. Clin. Exp. Allergy 45, 1126–1137 (2015).
Wang, L. L. et al. CD39/CD73 and the imbalance of Th17 cells and regulatory T cells in allergic asthma. Mol. Med. Rep. 8, 1432–1438 (2013).
Fang, X., Chen, Z. & Tang, R. Peripheral pathological type 2 innate lymphoid cells associate with human asthma control status. Am. J. Respir. Crit. Care Med. 193, A7560–A7560 (2016).
Muehling, L. M. & Lawrence, M. G. & Woodfolk, J. A. Pathogenic CD4+ T cells in patients with asthma.J. Allergy Clin. Immunol. 140, 1523–1540 (2017).
Fahy, J. V. Eosinophilic and neutrophilic inflammation in asthma: insights from clinical studies. Proc. Am. Thorac. Soc. 6, 256–259 (2009).
Mjosberg, J. & Eidsmo, L. Update on innate lymphoid cells in atopic and non-atopic inflammation in the airways and skin. Clin. Exp. Allergy 44, 1033–1043 (2014).
Mjosberg, J. M. et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12, 1055–1062 (2011).
Pulendran, B. & Artis, D. New paradigms in type 2 immunity. Science 337, 431–435 (2012).
Shih, H. Y. et al. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell 165, 1120–1133 (2016).
Hondowicz, B. D. et al. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity 44, 155–166 (2016).
He, R., Oyoshi, M. K., Jin, H. & Geha, R. S. Epicutaneous antigen exposure induces a Th17 response that drives airway inflammation after inhalation challenge. Proc. Natl Acad. Sci. USA 104, 15817–15822 (2007).
Wilson, R. H. et al. Allergic sensitization through the airway primes Th17-dependent neutrophilia and airway hyperresponsiveness. Am. J. Respir. Crit. Care Med. 180, 720–730 (2009).
Ano, S. et al. Transcription factors GATA-3 and RORgammat are important for determining the phenotype of allergic airway inflammation in a murine model of asthma. J. Immunol. 190, 1056–1065 (2013). (Baltimore, Md: 1950).
Choy, D. F. et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci. Transl. Med. 7, 301ra129 (2015).
Hamzaoui, A. et al. Transcriptional characteristics of CD4 T cells in young asthmatic children: RORC and FOXP3 axis. J. Inflamm. Res. 4, 139–146 (2011).
Zhao, Y., Yang, J., Gao, Y. D. & Guo, W. Th17 immunity in patients with allergic asthma. Int. Arch. Allergy Immunol. 151, 297–307 (2010).
Kanellopoulou, C. & Muljo, S. A. Fine-tuning Th17 cells: to be or not to be pathogenic? Immunity 44, 1241–1243 (2016).
Van Dyken, S. J. et al. A tissue checkpoint regulates type 2 immunity. Nat. Immunol. 17, 1381–1387 (2016).
Chae, W. J. et al. The Wnt antagonist dickkopf-1 promotes pathological type 2 cell-mediated inflammation. Immunity 44, 246–258 (2016).
Ramesh, R. et al. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med. 211, 89–104 (2014).
Szklarczyk, D. et al. STRINGv10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015). (Database issue).
Snel, B., Lehmann, G., Bork, P. & Huynen, M. A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444 (2000).
Becht, E. et al. Evaluation of UMAP as an alternative to t-SNE for single-cell data. doi: https://doi.org/10.1101/298430. bioRχiv, (2018)
Rossella Melchiotti, Kia Joo Puan, Anand Kumar Andiappan, Tuang Yeow Poh, Mireille Starke, Li Zhuang, Kerstin Petsch, Tuck Siong Lai, Fook Tim Chew, Anis Larbi, De Yun Wang, Michael Poidinger, Olaf Rotzschke, Genetic analysis of an allergic rhinitis cohort reveals an intercellular epistasis between FAM134B and CD39. BMC Medical Genetics 15:73 (2014)
Acknowledgements
We gratefully acknowledge the Flow Cytometry and Immunomonitoring Platform of Singapore Immunology Network (SIgN). Anis Larbi is a Marylou Ingram Scholar (International Society for Advancement of Cytometry). We thank Dr. Glenn Wong (Singapore Immunology Network) and Marie Clare Strickland (University of Southampton) for their assistance in editing the manuscript. This study was supported by National Science Funds for Young Scientist (81600018) provided by the National Natural Science Foundation of China (NSFC).
Author information
Authors and Affiliations
Contributions
Y.L. and H.K. designed the experiments, performed statistical analysis, and wrote the manuscript. Y.L., H.K. and S.W.T. performed the experiments. H.K., A.L., T.P.N. and H.B. conceived this study, supervised the experiments, and reviewed the manuscript. E.B. and E.W.N. generated the customised UMAP script for the unbiased analysis of flow cytometry data.The authors are responsible for the writing and the content of this article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lu, Y., Kared, H., Tan, S.W. et al. Dynamics of helper CD4 T cells during acute and stable allergic asthma. Mucosal Immunol 11, 1640–1652 (2018). https://doi.org/10.1038/s41385-018-0057-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0057-9
This article is cited by
-
Lactobacillus rhamnosus Modulates Lung Inflammation and Mitigates Gut Dysbiosis in a Murine Model of Asthma-COPD Overlap Syndrome
Probiotics and Antimicrobial Proteins (2023)
-
The Emerging Roles of T Helper Cell Subsets and Cytokines in Severe Neutrophilic Asthma
Inflammation (2022)