Abstract
Dietary antigen acquisition by lamina propria (LP) dendritic cells (DCs) is crucial to induce oral tolerance and maintain homeostasis. However, encountering innocuous antigens during infection can lead to inflammatory responses, suggesting processes may limit steady-state luminal antigen capture during infection. We observed that goblet cell (GC) associated antigen passages (GAPs), a steady-state pathway delivering luminal antigens to LP-DCs, are inhibited during Salmonella infection. GAP inhibition was mediated by IL-1β. Infection abrogated luminal antigen delivery and antigen-specific T cell proliferation in the mesenteric lymph node (MLN). Antigen-specific T cell proliferation to dietary antigen was restored by overriding GAP suppression; however, this did not restore regulatory T cell induction, but induced inflammatory T cell responses. Salmonella translocation to the MLN required GCs and correlated with GAPs. Genetic manipulations overriding GAP suppression, or antibiotics inducing colonic GAPs, but not antibiotics that do not, increased dissemination and worsened outcomes independent of luminal pathogen burden. Thus, steady-state sampling pathways are suppressed during infection to prevent responses to dietary antigens, limit pathogen entry, and lessen the disease. Moreover, antibiotics may worsen Salmonella infection by means beyond blunting gut microbiota colonization resistance, providing new insight into how precedent antibiotic use aggravates enteric infection.
Similar content being viewed by others
Introduction
The single-layer epithelium lining the gastrointestinal tract is exposed to a wide variety of substances, ranging from food and commensal microbes to enteric pathogens. In the steady state, the immune system underlying this epithelium samples the luminal contents to promote tolerance to dietary antigens,1,2,3 a process that is central to maintaining immune homeostasis and health. However, during enteric infection, the immune responses change to promote immunity and pathogen clearance. This shift in immune phenotype is not dictated solely by the nature of the antigens to which the immune system is responding, as evidenced by the induction of inflammatory responses to commensal gut flora during enteric infection.4 Thus, exposure to innocuous luminal substances during enteric infection can lead to inappropriate inflammatory responses; however, these responses are not commonly observed. This indicates that mechanisms could exist to limit immune exposures to innocuous luminal antigens during infection. Moreover, pathogens might co-opt the pathways of steady-state immune sampling and use them as a portal to cross the epithelium, providing an additional impetus for the host to regulate steady-state antigen sampling processes during enteric infection. However, whether steady-state luminal antigen acquisition pathways are suppressed during enteric infection and the implications of these events on the course of enteric infection are largely unexplored.
Luminal substances can traverse the epithelium by several pathways including paracellular leak, epithelial barrier breach, transcytosis by M cells, passage through goblet cells (GCs), and direct capture by lamina propria (LP) dendritic cells (DCs).5,6,7,8,9,10 Of these, transfer via GCs, or GC-associated antigen passages (GAPs), represents a major pathway for steady-state luminal antigen transfer to the LP-DCs in a manner capable of inducing antigen-specific T cell responses.7 Moreover, GAP formation is a controlled process, which can limit inappropriate exposure of the immune system to luminal substances.11,12,13 In addition, enteric pathogenic bacteria can use GCs, and commensal bacteria can use inappropriately formed colonic GAPs, to cross the epithelium.12,13,14 Together these observations suggest that inhibiting GAPs could limit antigen-specific T cell responses to dietary antigens and pathogen translocation during infection, and therefore might represent a physiologic response to enteric infection. Accordingly, we investigated how GAPs and antigen-specific T cell responses toward dietary antigen are altered during infection with Salmonella enterica subspecies I serovar Typhimurium.
Here, we report that infection with Salmonella inhibits GAPs, a steady-state luminal antigen acquisition pathway. Inhibition of GAPs during infection prevented inflammatory T cell responses to dietary antigen. In addition, we observed that translocation of Salmonella to the mesenteric lymph node (MLN) required GCs and correlated with the presence of GAPs. Pretreatment with antibiotics has a well-described effect in abrogating colonization resistance by the gut microbiota and potentiating disease by allowing Salmonella to expand in the gut lumen.15,16,17,18,19,20,21 Recently it has also been demonstrated that the dysbiosis induced by antibiotic pretreatment allows colonic GAP formation and translocation of commensal gut bacteria.11,12,13 We also observed that manipulations overriding GAP suppression, including antibiotic pretreatment, facilitated the dissemination of Salmonella, worsened disease course, and reduced survival, independent of overcoming colonization resistance of the gut microbiota. Thus, inhibition of this steady-state luminal antigen acquisition pathway during enteric infection serves to limit inappropriate antigen-specific T cell responses to dietary antigens and to limit pathogen translocation. Moreover, we describe an unappreciated effect of antibiotics potentiating and worsening enteric infection that is distinct from overcoming colonization resistance of the gut microbiota.
Results
Salmonella inhibits small intestinal GAPs in a cell intrinsic manner
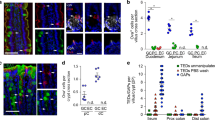
To evaluate if GAPs were inhibited during enteric infection, mice were given 5 × 108 CFU wildtype or invasion-deficient (ΔinvG) Salmonella into the small intestinal (SI) lumen, and SI GAPs density measured 1 h later. Wildtype Salmonella, and to a lesser extent ΔinvG Salmonella, acutely suppressed SI GAP formation (Fig. 1a, b). Further, we observed that SI GAPs were suppressed 1 day following oral gavage of 5 × 107 CFU wildtype Salmonella, and to lesser extent, ΔinvG Salmonella (Supplementary Fig. S1a). GAP inhibition was even more prominent 2 and 3 days after infection (Fig. 1b and Supplementary Fig. 1a). While GC numbers decreased at day 3 of infection, the reduction in GAPs was more pronounced than the decrease in GCs (Supplementary Fig. 1b), and therefore the reduction in GAPs could not be explained by GC loss alone. Thus, Salmonella inhibits SI GAPs for days following infection.
Goblet cell-associated antigen passage (GAP) formation is inhibited by Salmonella in a MyD88 and EGFR dependent manner. a Fluorescent images of SI villus cross section of C57BL/6 mice given luminal 10 kD dextran (red) and DAPI (blue), 1h after receiving luminal PBS (left), 5 × 108 CFU invasion-deficient Salmonella (ΔinvG Sal; center) or 5 × 108 CFU wildtype Salmonella (WT-Sal; right). b Density of GAPs in C57BL/6 mice, given 5 × 108 CFU or wildtype or ΔinvG Salmonella in the SI lumen 1 hour (hr) earlier (left side) or given 5 × 107 CFU wildtype or ΔinvG Salmonella orally 2 days earlier (right side). c MyD88fl/fl Math1Cre*PR and d EGFRf/f Math1Cre*PR mice and littermate controls were treated with RU486 to delete MyD88 or EGFR from GCs and were administered with 5 × 108 CFU of wildtype Salmonella or PBS for 1 h within the SI lumen (left side), or gavaged with 5 × 107 CFU wildtype Salmonella and SI tissue sections were evaluated 2 days later (right side). Graphs depict the density of GAPs in SI villus cross section of uninfected and infected mice. Data presented as the mean ± SEM. Scale bar in a = 50 μm; *p < 0.05, ns = not significant, n = 5 or more mice with 60 or more villus cross sections per mouse examined for each condition
GAP formation is induced by acetylcholine (ACh) acting on the muscarinic ACh receptor 4 (mAChR4) expressed by GCs.11 Responsiveness to ACh by GCs to form GAPs is inhibited by the activation of the epidermal growth factor receptor (EGFR) in GCs.11 Inhibition of GAP formation can occur via activation of EGFR directly in GCs by luminal EGF or by GC intrinsic MyD88-dependent sensing of the luminal microbiota in colonic GCs, resulting in EGFR trans-activation.11 However, GC intrinsic MyD88-dependent sensing of the microbiota does not suppress SI GAP formation.11 We explored if these pathways previously identified to inhibit GAPs were required for GAP suppression during acute infection with Salmonella. Inhibition of EGFR activation (EGFRi) in the absence of infection did not increase SI GAPs (Supplementary Fig. 2a), consistent with prior observations that EGFR is not activated in SI GCs and not suppressing SI GAP formation in the steady state.11 However, pharmacologic inhibition of EGFR activation 2 days after Salmonella infection partially reversed the suppression of SI GAPs (Supplementary Fig. 2a). The inability to completely reverse GAP inhibition may be due to the short half-life of this pharmacologic inhibitor of EGFR activation.22 Therefore, we evaluated GAPs in mice where EGFR or Myd88, the upstream inducer of EGFR trans-activation, was deleted in GCs. Mouse atonal homologue 1 (Math1) is a transcription factor necessary for the development of neurons23,24 and secretory intestinal epithelial lineages including GCs, Paneth cells, and enteroendocrine cells. 25 Previous studies have demonstrated that mice with Cre recombinase inserted into the Math1 locus effectively targets intestinal GCs.11,24 We found that GC deletion of EGFR, or its upstream activator MyD88, using the inducible Math1-driven Cre recombinase,11,12,24 completely reversed SI GAPs inhibition 1 h after luminal administration of Salmonella as well as 2 days following oral infection, but did not increase SI GAPs in the absence of infection (Fig. 1c, d). Thus, inhibition of SI GAPs during Salmonella infection occurs by activation of MyD88- and EGFR-dependent signaling pathways previously identified to inhibit colonic GAPs in the steady state.11
IL-1β inhibits GAPs during Salmonella infection
Heat-killed cecal contents do not inhibit SI GAPs in the steady state, and deletion of MyD88 in GCs does not augment SI GAP density (Fig. 1c and ref.11). This may be due to lower TLR expression and higher expression of inhibitors of TLR signaling by SI GCs when compared with colonic GCs.11 These observations indicate that microbial products do not suppress SI GAPs via MyD88 signaling in GCs in the steady state. Similarly, we observed that heat-killed Salmonella did not inhibit SI GAPs (Supplementary Fig. 2b). However, GAP inhibition during Salmonella infection was dependent upon MyD88 (Fig. 1c), suggesting that other stimuli may be activating MyD88 during infection. Further, we observed that SI GAP inhibition during Salmonella infection was associated with increased phosphorylation of EGFR in the epithelium, but did not correlate with increased levels of EGF (Fig. 2a, b), prompting us to explore other MyD88-dependent pathways leading to EGFR activation. Salmonella infection induces IL-1β production by stromal cells26 and mononuclear phagocytes,27 and IL-1β can signal via MyD88,28 which activates the EGFR in GCs.11 We found that IL-1β was significantly elevated in the epithelium 2 days following Salmonella infection. Wildtype Salmonella induced significantly more IL-1β than ΔinvG Salmonella (Fig. 2c), correlating with increased GAP inhibition by wildtype Salmonella (Fig. 1b). Moreover, administration of recombinant IL-1β systemically or into the gut lumen rapidly inhibited SI GAPs (Fig. 2d), and inhibiting active IL-β release with the pan-caspase inhibitor Z-VAD-FMK reversed the inhibition of SI GAPs following Salmonella infection (Fig. 2e). While this supports the involvement of IL-1β in GAP inhibition, Z-VAD-FMK may have effects beyond suppressing active IL-1β release. However, we observed that germline deletion of IL-1 receptor 1 (Il1r1−/−) did not alter SI GAP density in the steady state, but reversed SI GAP inhibition 2 days following oral Salmonella infection as well as 1 h following luminal Salmonella administration (Fig. 2f), thus confirming the necessity of this pathway for the suppression of SI GAPs during infection.
IL-1β inhibits SI GAPs during Salmonella infection. Enzyme-linked immunosorbent assays for a phospho-EGFR, b EGF, and c IL-1β in SI epithelium from uninfected C57BL/6 mice or C57BL/6 mice infected with 5 × 107 CFU ΔinvG or wildtype Salmonella 2 days earlier. d Density of GAPs in C57BL/6 mice 1 h after i.p. injection or luminal injection of vehicle or 100 ng recombinant IL-1β. e Density of SI GAPs in C57BL/6 mice 2 days after oral PBS or 5 × 107 wildtype Salmonella and daily injection with vehicle or 10 µg/kg of pan-caspase inhibitor (Z-VAD-FMK). f Density of GAPs in C57BL/6 or IL1r−/− mice, given 5 × 108 CFU of wildtype Salmonella in the SI lumen 1 hr earlier (left panel) or given 5 × 107 CFU wildtype Salmonella orally 2 days earlier. Data presented as the mean ± SEM; *p < 0.05, ns = not significant, n = 5 or more mice with 60 or more villi cross sections evaluated for each condition.
Suppression of SI GAPs during Salmonella infection limits responses to dietary antigen in draining MLN
In the steady state, inhibition of GAPs or deletion of GCs/GAPs abrogates the ability of LP-DCs to acquire luminal antigen in a manner capable of inducing adaptive immune responses,7,11 a process that is critical for the induction of T cell responses in the draining MLN.1,2 This suggests that GAP inhibition during enteric infection would suppress antigen-specific T cell responses to orally administered antigens. In addition, Salmonella infection can inhibit antigen presentation by DCs29 and/or kill DCs,29,30 further impairing generation of immune responses to dietary antigen. Conversely, Salmonella infection increases epithelial barrier permeability31 and induces LP-DC extension of trans-epithelial dendrites (TED) into the lumen,9,32 both of which might increase luminal antigen capture and antigen-specific T cell responses to dietary antigens. To determine how Salmonella infection influences T cell responses to dietary antigen, uninfected and infected mice were injected i.v. with CFSE-labeled ovalbumin (Ova)-specific splenic T cells isolated from OTII T cell receptor transgenic mice, gavaged with Ova, and T cell proliferation in the MLN was quantified 48 h later. Salmonella-infected mice had significantly suppressed antigen-specific T cell proliferation to luminal Ova in the MLN (Fig. 3a, b), despite equivalent abilities of OTII T cells to traffic to the MLN and despite having equivalent ability to respond to systemic Ova (Supplementary Fig. 3a and b). This suggests that the increased gut permeability and LP-DC TED extension during Salmonella infection do not translate into enhanced immune responses to luminal antigens and that the defect in antigen-specific T cell proliferation in the MLN to luminal Ova is in part due to lack of antigen delivery to the MLN during infection. Further supporting this, we observed that inhibition of EGFR activation, which relieves GAP inhibition during Salmonella infection (Supplementary Fig. 2), reversed the impairment of antigen-specific T cell proliferation to dietary antigen during infection, but did not increase proliferation in the absence of infection (Fig. 3a, b). Moreover, the impaired T cell proliferation to dietary antigen seen during infection was reversed by GC-specific deletion of MyD88 (Fig. 3c, d), which relieves the suppression of GAPs during Salmonella infection (Fig. 1c). A significant proportion of the OTII T cells in the MLN of uninfected Ova-gavaged mice expressed Foxp3, indicating differentiation into Tregs (Fig. 3e, f). In contrast, Salmonella infection significantly reduced the population of OTII T cells expressing Foxp3 in the draining MLN after Ova gavage (Fig. 3e, f). Notably, while GC-specific deletion of MyD88 restored antigen-specific T cell proliferation to dietary Ova during Salmonella infection (Fig. 3c), this did not restore Foxp3 expression (Fig. 3e, f), but did induce an increase in the number of IL-17-producing OTII T cells responding to dietary Ova (Fig. 3g, h). In light of studies demonstrating that inflammatory responses are generated toward commensal bacterial antigens during infection,4 our findings suggest that one outcome of inhibiting GAPs during infection is to limit inflammatory antigen-specific T cell responses to innocuous dietary substances in the gut lumen.
Inhibition of GAP formation by Salmonella impairs antigen-specific T cell proliferation and Treg induction to dietary antigen in the draining MLN. a Histograms and b quantification of proliferation of adoptively transferred CSFE labeled Ova-specific CD4+ OTII T cells in the MLN of C57BL/6 mice that were uninfected or infected with 5 × 107 CFU Salmonella and gavaged with Ova. c Flow cytometry plots and d quantification of proliferation of adoptively transferred CFSE labeled Ova-specific CD4+ OTII T cells in the MLN of mice lacking MyD88 in GCs or their littermate controls that were uninfected or infected with 5 × 107 CFU Salmonella adoptively transferred OTII T cells one day later, gavaged with Ova 2 days after infection and analyzed after 2 days. Mice were treated with RU486 daily starting at the time of infection until Ova gavage. e, g Flow cytometry plots and f, h quantification of Foxp3 and IL-17 expression by adoptively transferred Ova-specific CD4+ OTII T cells in the MLN of mice lacking MyD88 in GCs or littermate controls that were uninfected or infected with 5 × 107 CFU Salmonella and gavaged with Ova. Data are pooled from two independent experiments, each with 6–8 mice per group; *p < 0.05, ns = not significant. Data presented as the mean ± SEM.
Salmonella uses GCs and GAPs as a portal of entry
Animal models and in vitro studies have identified several strategies Salmonella can employ to cross the epithelial barrier, including barrier breach, transcytosis of M cells or other epithelial cells, direct capture by LP-DC extension of dendrites into the lumen, and paracellular penetration between epithelial cells.10,33 In addition to these pathways, the enteric pathogen Listeria monocytogenes has been found to selectively target GCs in order to cross the SI epithelial barrier.14 Moreover, translocation of gut commensals across colonic epithelium requires GCs and is associated with the formation of colonic GAPs.12 These observations prompted us to evaluate if Salmonella might use GCs and GAPs in the SI as a portal for entry. We observed that 2 h following intraluminal inoculation, Salmonella preferentially localized with, and could be found within, SI GCs that had formed GAPs (Fig. 4a). The SI and colonic lymphatics largely drain into distinct MLNs, and evaluation of the MLNs draining the SI and colon independently can inform where bacteria translocate across the epithelium in the gut.12,13,34 Two days following oral infection, wildtype Salmonella was nearly absent from colon-draining MLN, but was easily found in SI-draining MLN (Fig. 4b), indicating that wildtype Salmonella predominantly traversed the SI epithelium. Deletion of GCs abrogated dissemination of Salmonella to the SI-draining MLN, but did not affect colonization of Peyer’s patches (PP) or spleen bacterial loads 2 days after infection (Fig. 4b). This indicates that GCs are required for Salmonella to cross the SI epithelium to access the draining MLN early in infection, but are not required for infection of the PP or dissemination to the spleen. This also suggests that Salmonella dissemination to the spleen might be independent of traversing the non-follicle bearing epithelium in the SI, which has been indicated by others.35
Dissemination of Salmonella to draining MLNs requires GCs. a Fluorescent images of an SI villus cross section (left) and a confocal image of an SI GAP (right) in C57BL/6 mice 2 hr after receiving luminal 10 kDa dextran (red) and GFP-expressing Salmonella (green); nuclear stain DAPI (blue). Colony forming units (CFUs) in the colon-draining MLN (C-MLN), SI-draining MLN (S-MLN), Peyer’s patches (PP), and spleen (SPL) 2 days after infection with 5 × 107 CFU wildtype Salmonella in b goblet cell-deficient (Math1f/f vil-Cre-ERT2) mice or littermate control (Math1f/f) mice, c untreated or streptomycin pretreated C57BL/6 mice or d goblet cell-deficient mice littermate controls pretreated with streptomycin. Data are pooled from three independent experiments, each with 5 mice per group; *p < 0.05, ns = not significant, scale bar = 50 μm. Data are presented as the mean ± SEM.
Oral streptomycin treatment of mice prior to Salmonella infection potentiates colitis and is used to model the pathogenesis of human Salmonella enterocolitis.17 Antibiotic pretreatment disrupts the gut microbiota and allows Salmonella to more effectively colonize the lumen, increasing colonic inflammation and enhancing dissemination.15,16,17,18,19,20,21 Moreover, antibiotic pretreatment allows ΔinvG Salmonella to traverse the epithelium and disseminate to MLN (Supplementary Fig. 4a and refs. 36,37). We found that streptomycin pretreatment augmented bacterial dissemination specifically to the colon-draining MLN and augmented bacterial burden in the spleen, without altering bacterial burden in the SI-draining MLN or PP (Fig. 4c). GAPs form spontaneously in the SI, but are suppressed in the colon due to GC-intrinsic microbial sensing, which inhibits GC responsiveness to ACh;11 however, colonic GAPs are induced following treatment with some antibiotics and can facilitate translocation of commensal bacteria.12,13 We observed that deletion of GCs in streptomycin-pretreated mice abrogated both the ability of Salmonella to disseminate to the colon- and SI-draining MLNs and the increase in spleen titers (Fig. 4d), indicating that GCs were required for Salmonella dissemination to the MLNs in the streptomycin pretreatment model.
In streptomycin-pretreated mice infected with wildtype Salmonella, the bacteria translocating to the colon-draining MLN included both Salmonella and gut commensal bacteria, while those translocating to the SI-draining MLN were Salmonella alone (Supplementary Fig. 4b). This is consistent with prior studies demonstrating that colonic GAPs, but not SI GAPs, facilitate the translocation of commensal bacteria12,13 and indicates that these properties of SI and colonic GAPs to translocate commensal bacteria do not change during infection. Pretreatment with kanamycin, which is also used in a Salmonella colitis model,38,39 similarly induced colonic GAPs and enhanced translocation to the colon-draining MLN and spleen, but not to the SI-draining MLN (Supplementary Fig. 4c and d). These observations suggest that during the early phase of infection, GCs are required for translocation of Salmonella across the colonic epithelium in the streptomycin or kanamycin pretreatment models and that the enhanced pathogen burden in the spleen following streptomycin or kanamycin pretreatment is in part due to increased translocation across the colonic epithelium via GCs/GAPs.
Overriding GAP suppression worsens the course of Salmonella infection
Prior antibiotic use has been associated with an increased risk of Salmonella infection.40,41,42 A single, clinically relevant dose of many, but not all, antibiotics induces colonic GAPs and commensal bacterial translocation to the colon-draining MLN in mice. Therefore, we evaluated the effects of pretreatment with lower doses of antibiotics that do (streptomycin and vancomycin) and do not (neomycin and tetracycline) induce GAPs12 on Salmonella infection. All antibiotic pretreatments significantly increased the luminal pathogen load when compared to mice not receiving antibiotics, with no significant differences in luminal Salmonella burden between the antibiotic treatment groups (Fig. 5a). Pretreatment with antibiotics that induce colonic GAPs, but not pretreatment with those that do not induce colonic GAPs, significantly increased dissemination to the colon-draining MLN and reduced survival (Fig. 5b, c). We observed that like spontaneously forming SI GAPs, colonic GAPs induced after antibiotic pretreatment were also inhibited 2 days after Salmonella infection (Supplementary Fig. 5a). Like SI GAPs, colonic GAPs induced following antibiotic administration were inhibited by IL-1β treatment in an EGFR-dependent manner (Supplementary Fig. 5b and c).
Pretreatment with oral antibiotics that induce colonic GAPs, but not with those that do not induce colonic GAPs, increases Salmonella dissemination and reduces survival. a Salmonella in the colon contents and b CFUs in the colon-draining MLN and c Kaplan–Meier survival curves in untreated C57BL/6 mice or C57BL/6 mice pretreated with neomycin (500 µg), tetracycline (500 µg), streptomycin (500 µg), or vancomycin (250 µg) 1 day prior to infection with 5 × 107 CFU Salmonella. Data represented as the mean ± SEM; *p < 0.05, ns = not significant, n = 5 or more mice.
While the above observations suggest that antibiotic pretreatment enables more severe Salmonella infection, and this can be independent of overcoming colonization resistance of the gut microbiota, antibiotics may have a number of effects enhancing dissemination and worsening infection that are unrelated to their ability to induce colonic GAPs. Therefore, we evaluated the effect of deletion of MyD88 or EGFR in GCs to override the suppression of GAPs during Salmonella infection independent of manipulations of the gut microbiota by antibiotics. EGFRfl/flMath1Cre*PR mice or MyD88fl/fl Math1Cre*PR mice and their littermate controls were treated with RU486 prior to and during the first 2 days of infection to open GAPs in a pattern seen after a single dose of antibiotics.12 Overriding the suppression of GAP formation via deletion of MyD88 or EGFR in GCs was sufficient to exacerbate Salmonella infection, as evidenced by increased production of inflammatory cytokines, increased colitis, and increased mortality when compared with littermate controls; indeed, mice lacking Myd88 or EGFR in GCs exhibited a disease course and survival similar to mice receiving streptomycin pretreatment (Fig. 6a–e). Interestingly, bacterial burden in the colonic MLN was similar among EGFRfl/flMath1Cre*PR, MyD88fl/fl Math1Cre*PR, and streptomycin-pretreated wildtype mice (Fig. 6e), despite the presence of a higher luminal pathogen burden in the streptomycin pretreatment group (Fig. 6f). Together, these data suggest that antibiotics may exacerbate Salmonella infection by inducing colonic GAPs and allowing increased pathogen dissemination.
Overriding GAP inhibition results in increased dissemination, worsened colitis, and reduced survival independent of alterations in luminal Salmonella burden. a Representative hematoxylin- and eosin-stained colon sections, b Kaplan–Meier survival curves, c histological scores, d serum cytokine concentrations, e CFUs in colon-draining MLNs, and f luminal Salmonella burden in untreated C57BL/6 mice, streptomycin-pretreated C57BL/6 mice, mice lacking EGFR in GCs (EGFRf/f Math1PR*Cre), mice lacking MyD88 in GCs (MyD88f/f Math1P*RCre), and floxed littermate controls infected with 5 × 107 CFU of Salmonella. Mice in a and c–f were analyzed 6 days following infection; n = 5 or more mice, *p < 0.05. Data presented as ±SEM. Scale bar = 100 µm
Discussion
The intestinal epithelium is a major interface between the host and the environment, which includes food, the commensal microbiota, and potential pathogens. The delivery of luminal antigens across the intestinal epithelium in a manner capable of inducing adaptive immunity is a critical and early step in intestinal immune responses to luminal substances. In the steady state, these responses are characterized by the induction of Tregs enforcing tolerance to dietary and commensal antigens.43,44,45 However, during infection, the tone of the immune response changes to promote the pathogen clearance and protective immunity. Inflammatory T cell responses can be induced to luminal non-pathogens encountered in the setting of enteric infection.4 These inflammatory T cell responses can be long-lived, and therefore limiting the immune system’s exposure to non-pathogenic antigens during infection would reduce the opportunity for developing inappropriate inflammatory responses. Here, we explored how an enteric infection alters the delivery of luminal antigens to immune system early in the course of infection. We focused our studies on GCs and GAPs, as the formation of GAPs is tightly controlled, and GAPs are a major pathway delivering antigens to the LP immune system in a manner capable of inducing adaptive immunity.7,11 Thus, the regulation of GAP formation is a potential mechanism limiting exposure to luminal substances during infection (Supplementary Fig. 6).
In the steady state, GAPs form continuously in the SI due to GC responsiveness to ACh.7,11 However, GC responsiveness to ACh is repressed in the colon, as MyD88-dependent sensing of the microbiota activates EGFR in GCs suppresses responses to ACh.11 Microbial sensing via MyD88 does not suppress SI GAP formation despite similar levels of expression of MyD88 in SI and colon GCs; however, the ability of EGFR activation to inhibit GAP formation in GCs is retained in the SI.11 This distinction may be related to lower expression of TLRs and higher expression of suppressors of TLR signaling in SI GCs when compared with colonic GCs,11 or other yet to be identified factors. In the present work, we show that infection with wildtype Salmonella acutely inhibited GAPs, and this inhibition was dependent upon MyD88 and EGFR in GCs. Like the TLR signaling pathway, IL-1 receptor signaling relies on MyD88.28 Infection with Salmonella rapidly induces IL-1β release by stromal cells26 and in intestinal phagocytes via activation of NLRC4-inflammasomes.46 NLRC4 activation and IL-1β release by intestinal phagocytes during enteric infection is dependent upon an intact type III secretion system,46,47 which is disrupted in ΔinvG Salmonella,48,49,50 providing an explanation for the higher efficiency of wildtype Salmonella at MyD88-dependent GAP inhibition than the ΔinvG mutant. Indeed, we found that IL-1β was significantly elevated following Salmonella infection and correlated with the ability of wildtype Salmonella to more effectively inhibit GAPs when compared with ΔinvG Salmonella, recombinant IL-1β could rapidly inhibit GAPs, and that Salmonella failed to inhibit GAPs in IL-1 receptor-deficient mice.
We observed that Salmonella infection inhibited dietary antigen-specific T cell proliferation and Foxp3 induction. Interestingly, Salmonella has been reported to induce increased permeability of the intestinal barrier31 and to promote the extension of TEDs by LP-DCs.9,32 Despite these reported effects, we observed that Salmonella inhibition of GAPs correlated with reduced immune response to dietary antigen during infection. Processes other than GAP inhibition can limit antigen-specific T cell proliferation to luminal substances during infection. When DCs are infected with Salmonella, presentation of peptides on MHCII and the ability to stimulate antigen-specific T cell proliferation is reduced.29 Further, Salmonella has been reported to kill DCs,30 which would limit their ability to induce antigen-specific T cell proliferation toward luminal substances. While this may be occurring in our studies, we do not believe it explains the observed reduction in T cell proliferation to luminal antigens in the draining MLN. Overriding GAP suppression by wildtype Salmonella, by deleting MyD88 in GCs, or by pharmacologically inhibiting EGFR signaling, relieved the inhibition of T cell proliferation to luminal antigen in the MLN. These maneuvers targeting GCs do not reduce the pathogen burden and, due to the ability of invasive Salmonella to use GAPs as a portal of entry, may instead increase the pathogen burden, while simultaneously increasing T cell proliferation in response to luminal antigen in the MLN. These data are inconsistent with reduced antigen presentation by infected DCs or killing of DCs by Salmonella as the only events suppressing T cell proliferation to luminal antigen during infection.
In the steady state, tolerance to dietary antigen occurs in the SI and is characterized by the induction of tolerogenic Foxp3+ Tregs.1,3,51 In accordance with this, we observed substantial induction of Foxp3+ Ova-specific T cells in the MLN of uninfected mice receiving dietary Ova. In line with our observations of reduced tolerance to dietary antigen during infection, we found that induction of Ova-specific Foxp3+ T cells was reduced during Salmonella infection. However, deletion of MyD88 to relieve inhibition of GAPs during infection did not restore Treg induction, despite restoring antigen-specific T cell proliferation to dietary Ova, and in fact induced more IL-17 producing Ova-specific T cells in the MLN. Thus, similar to T cell responses to commensal bacterial antigens,4 inflammatory responses are also generated in response to dietary antigens encountered during infection.
We found that wildtype Salmonella uses GCs/GAPs to cross the SI epithelium and gain access to the draining MLN. This was the predominant pathway delivering Salmonella to the MLN early in infection, as evidenced by the near absence of Salmonella in the MLN following deletion of GCs. Moreover, we found that overriding the suppression of GAPs during Salmonella infection increased the pathogen burden in the MLN indicating that inhibition of GAPs may be a mechanism limiting pathogen dissemination. While it appears paradoxical that Salmonella would both inhibit GAPs and use them as a route of entry, we believe the pathogens found in the MLN are early invaders, some of which induce IL-1β release by LP mononuclear phagocytes subsequently inhibiting GAPs and limiting further dissemination. One limitation of our studies is that they were largely focused on early time points in infection, raising the possibility that other routes of entry may contribute to dissemenation to the MLN at later time points. While it is possible that GAP inhibition during infection diminishes the host’s immune response to Salmonella, we observed that overriding GAP inhibition resulted in worsened disease, indicating that GAP inhibition during infection overall is favorable to the host and a physiologic response during infection. Potentially consistent with our findings, IL1r1−/− mice are more susceptible to orogastric infection, but not peritoneal infection by Salmonella,27 suggesting that loss of GAP inhibition by IL-1β and enhanced dissemination may contribute to this differential susceptibility to these routes of Salmonella infection.
Prior antibiotic use has been associated with an increased risk for infection by Salmonella in humans.40,41,42 The effect of antibiotics to increase the risk of, and potentiate, enteric infection may be due to a reduction of the mucus layer or alterations in the environment, supporting pathogen residence and growth.15,20,52 Indeed, elegant studies have demonstrated that disruption of the microbiota by antibiotics not only creates a space for Salmonella to colonize but also alters the nutrient environment to favor growth of Salmonella.15,20 Our observations indicate that the effect of antibiotics also extends to promoting Salmonella’s translocation across the colonic epithelium and dissemination. We observed a similarly increased disease severity in streptomycin-pretreated mice and in mice with cell type-specific deletion of components of the pathways normally suppressing GAP formation. This indicates that facilitating Salmonella translocation and dissemination, without disrupting the gut microbiota and increasing luminal pathogen load via antibiotic administration, was sufficient to potentiate disease. Further, these effects were seen in mice treated with antibiotics that do, but not in mice treated with antibiotics that do not, induce colonic GAPs, despite similar burdens of luminal Salmonella. Importantly, these observations may inform the design of therapeutic antimicrobial regimens to target enteric pathogens without facilitating their translocation.
In total, we demonstrate how the delivery of luminal substances to the intestinal immune system is altered early in the course of infection and how a steady-state antigen delivery pathway is used by an enteric pathogen as a portal of entry. We acknowledge that these studies were performed in an in vivo infection model and accordingly are complex and open to other interpretations regarding the events controlling antigen-specific T cell responses to dietary antigen and pathogen dissemination. However, combined with previous work demonstrating a role for GAPs in luminal antigen delivery and bacterial translocation, these findings indicate a critical role for GAP regulation limiting T cell responses to luminal antigens and pathogen dissemination during infection. Beyond this, we uncovered an unappreciated link between precedent antibiotic therapy and increased risk for enteric infection that is independent of the role of antibiotics overcoming colonization resistance of the gut microbiota (Supplementary Fig. 6). These observations provide a conceptual framework to better understand how controlled access to luminal antigens is integrated to promote appropriate mucosal immune responses in the steady state and during enteric infection.
Materials and methods
Mice
Mice used in this study were on the C57BL/6 background and bred in house. Where possible, cohoused littermates were used as experimental controls. C57BL/6 mice, IL1r1−/− mice,53 Myd88fl/fl mice,54 Math1Cre*PR mice,24 Math1fl/fl mice,25 OTII T cell receptor transgenic mice,55 congenic CD45.1 B6/SJL mice, and Foxp3GFP mice56 were initially purchased from The Jackson Laboratory (Bar Harbor, ME). CD45.1 B6/SJL mice, Foxp3GFP mice, and OTII mice were crossed to generate CD45.1 Foxp3GFP OTII mice for adoptive transfer of Ova-specific T cells. Mice with a Cre recombinase inserted into the Math1 locus, Math1Cre*PR mice, can effectively delete floxed genes or label secretory epithelial lineages, including GCs, Paneth cells, and enteroendocrine cells.11 Transgenic mice in which a tamoxifen-dependent Cre recombinase is expressed under the control of the villin promoter (vil-Cre-ERT2)57 mice were a gift from Sylvie Robine (Institut Curie, Paris, France). Math1fl/flvil-Cre-ERT2 mice and the injection protocol to induce GC deletion were previously described.11 EGFRfl/fl mice58 were a gift from Dr. David Threadgill, University of North Carolina. EGFRfl/fl mice and Myd88fl/fl mice were bred to Math1Cre*PR mice24 to generate mice with an inducible deletion of EGFR in GCs. To induce deletion of EGFR or MyD88, EGFRfl/flMath1Cre*PR mice or MyD88fl/fl Math1Cre*PR mice were treated with 200 µg RU486 (mifepristone, Caymen Chemicals, Ann Arbor, MI) dissolved in sesame oil (Acros Organics, Sigma-Aldrich) daily. All mice received regular chow diet and autoclaved water. Animal procedures and protocols were performed with the approval of the Institutional Animal Care and Use Committee at Washington University School of Medicine.
Bacteria strains/infection
Salmonella typhimurium wildtype strain SB300A1,59 its isogenic mutant ΔinvG, and S. typhimurium strain χ3761,60 transformed with a lethal balanced GFP expression plasmid to create Salmonella-GFP (unpublished strain created by Xin Zhang in the Miller lab) were grown with shaking overnight at 37 °C in Luria-Bertani (LB) broth, subcultured for 4 h, and washed twice with cold PBS prior to use.
Intraluminal infection was performed by innoculating 5 × 108 CFU of bacteria into the SI lumen of anesthetized mice. For oral Salmonella infections, mice were deprived of food for 4 h and gavaged orally with 5 × 107 CFU of bacteria in 200 μl PBS. Mice were kept without food and water for 1 h after infection. The final infection dose was verified by plating serial dilutions of the bacterial suspension on LB agar plates. In some experiments, mice were treated with 7.5 mg of streptomycin,61 or 10 mg kanamycin38,39 in 100 μl of PBS. To compare the effect of clinically relevant doses of different antibiotics, mice were gavaged with 500 µg neomycin, 500 µg tetracycline, 500 µg streptomycin, or 250 µg vancomycin 24 h prior to infection. For studies in figure 6, EGFRfl/flMath1Cre*PR mice and MyD88fl/fl Math1Cre*PR mice were treated with RU486 starting 5 days prior to infection and continuing for 2 days after infection to open colonic GAPs in a pattern similar to that seen after a single dose of antibiotics.12
Organ isolation and homogenization
MLN, spleen, and PP were retrieved and placed in gentamicin solution for 15 min. The gentamicin was washed away with cold PBS prior to grinding. All tissues were placed in 500 µl PBS and homogenized with a PowerGen 125 tissue homogenizer (Fisher Scientific). Live bacterial burden was determined by plating LB agar. Individual bacterial species were identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (Biotyper, Bruker Corporation, CA), as described previously.12
Enumeration of GAPs and GCs
SI and colonic GAPs were enumerated on fixed tissue sections as previously described. 7,11Briefly, tetramethylrhodamine-labeled 10 kD dextran was administered in the jejunum and proximal colon of anesthetized mice. After 1 h, mice were sacrificed and tissues thoroughly washed with cold PBS before fixing in 10% formalin buffered solution. Tissues were embedded in optimal cutting temperature compound (Fisher Scientific, Pittsburgh, PA); 7-μm sections were prepared, stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich), and imaged using an Axioskop 2 microscope (Carl Zeiss Microscopy, Thornwood, NY). GAPs were identified as dextran-filled columns measuring approximately 20 µm (height) × 5 µm (diameter) traversing the epithelium and containing a nucleus, and were enumerated as GAPs/villus cross section in the SI or GAPs/crypt cross section in the colon. Fixed SI tissue sections were stained with periodic acid-Schiff (PAS; Sigma-Aldrich) according to manufacturer’s instructions, and images were acquired on the Axioskop 2. In some experiments, mice were treated with 500 µg/kg tryphostin AG1478 (inhibitor of EGFR phosphorylation; Sigma-Aldrich) intraperitoneally (i.p.) 30 min prior to luminal dextran administration.
IL-1β and caspase inhibitor administration
Mice were treated i.p. or luminally with 100 ng recombinant IL-1β (R&D Systems) 1 h prior to intraluminal dextran administration. To study the effect of IL-1β signaling blockade, mice were administered i.p. with 10 mg/kg of pan-caspase inhibitor Z-VAD-FMK (R&D Systems) 1 h prior to intraluminal dextran administration.
Enzyme-linked immunosorbent assays
Serum levels of IL-6 and IFN-γ were measured using Mouse IL-6 and IFN-γ ELISA kit (eBioscience), according to manufacturer’s protocol. To measure the concentration of EGF (R&D Systems), phosphorylated-EGFR (R&D Systems) or IL-1β (eBioscience) in the SI-epithelium, a 5-cm portion of jejunum was opened longitudinally and washed with PBS. The epithelial cell layer was scraped from the submucosa with a glass slide, and cells were homogenized in 1 ml PBS. Cells were centrifuged, and supernatant was collected and ELISA performed per the manufacturer’s protocol.
Adoptive T cell transfer
To evaluate the effects of Salmonella infection on T cell trafficking and antigen-specific T cell proliferation, single-cell suspensions of Ova-specific T cells were prepared from spleens and MLNs of CD45.1+ OTII T cell receptor transgenic mice, and CD4 T cell enrichment was performed using magnetic beads (Stemcell Technology #19752, Vancouver, BC). Enriched CD4+ T cells were labeled with 2 µM CFSE (Invitrogen); 2 × 106 CFSE-labeled cells were adoptively transferred i.v. into sex-matched recipient mice 1 day after mock infection or infection with Salmonella. Twenty-four hours after transfer, mice were orally gavaged with 20 mg ovalbumin (Sigma) or in some experiments given 200 µg of i.v. ovalbumin, and after 2 days MLNs were removed and single-cell suspensions were prepared and analyzed by flow cytometry for CD45.1, CD3, Vβ5, Vα2, and CSFE. In experiments evaluating the trafficking of adoptively transferred T cells, mice were not gavaged with Ova, and transferred T cells evaluated on the same schedule.
To evaluate the phenotype of transferred cells, single-cell suspensions from spleen and MLNs from Ova-specific CD45.1+ Foxp3GFP OTII T cell receptor transgenic mice were flow cytometrically sorted for GFP−, Vβ5+, Vα2+, CD45.1+, CD62hi cells; 5 × 105 cells were intravenously administered into recipient (CD45.2) mice 1 day after infection. One day later, mice were gavaged with 20 mg OVA, 3 days later single-cell suspensions were isolated from the MLN, and evaluated for Foxp3 expression or were stimulated for 3 h with 10 ng/ml PMA, 500 ng/ml inomycin (both Sigma-Aldrich), and 2 µM Brefeldin A (eBioscience) to evaluate the cytokine expression by intracellular flow cytometry. Samples were acquired on a FACScan cytometer (BD Bioscience, San Jose, CA), and data analysis was performed using FlowJo software (TreeStar, Inc.).
Bacterial DNA isolation and quantification of Salmonella in intestinal contents
Entire SI and colonic contents were collected, placed in 1 mL lysis buffer and 200 mg 0.1 mm diameter zirconia silica beads (BioSpec, Bartlesville, OK) and vortexed on a bead beater (FastPrep 24, MP Biomedicals). DNA isolation was performed using DNA/RNA Extraction kit (Qiagen, Valencia, CA). Quantification of Salmonella was performed by real-time PCR using primers specific to siiA (ACGACTGGGATATGAACGGGGAA and TCGTTGTACTTGATGCTGCGGAG)62 and measured against a standard curve.
Histopathologic evaluation
On day 5 of Salmonella infection, colonic tissue was fixed in formalin and washed in ethanol before embedding. Hematoxylin and eosin staining was performed, and pathology was scored as previously described.12 In brief, edema (0–3 points), lymphocyte infiltration (0–3 points), and epithelial damage (0–3 points) were evaluated, and a total score between 0 and 9 points was obtained.
Statistical analysis
Data analysis was performed using GraphPad Prism (GraphPad Software Inc., San Diego, CA). Statistical significance was defined as p<0.05.
References
Worbs, T. et al Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J. Exp. Med 203, 519–527 (2006).
Schulz, O. et al Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med 206, 3101–3114 (2009).
Hadis, U. et al Intestinal tolerance requires gut homing and expansion of FoxP3+regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011).
Hand, T. W. et al Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556 (2012).
Turner, J. R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 9, 799–809 (2009).
Jang, M. H. et al Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proc. Natl Acad. Sci. USA 101, 6110–6115 (2004).
McDole, J. R. et al Goblet cells deliver luminal antigen to CD103+dendritic cells in the small intestine. Nature 483, 345–349 (2012).
Rescigno, M. et al Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367 (2001).
Niess, J. H. et al CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258 (2005).
Farache, J. et al Luminal bacteria recruit CD103+dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595 (2013).
Knoop, K. A., McDonald, K. G., McCrate, S., McDole, J. R. & Newberry, R. D. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 8, 198–210 (2015).
Knoop, K. A., McDonald, K. G., Kulkarni, D. H. & Newberry, R. D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 65, 1100–U1160 (2016).
Knoop, K. A. et al. Antibiotics promote the sampling of luminal antigens and bacteria via colonic goblet cell associated antigen passages. Gut Microbes, 1–12. https://doi.org/10.1080/19490976.2017.1299846 (2017).
Nikitas, G. et al Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med 208, 2263–2277 (2011).
Faber, F. et al Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534, 697–699 (2016).
Barthel, M. et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858 (2003).
Hapfelmeier, S. & Hardt, W. D. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol. 13, 497–503 (2005).
Croswell, A., Amir, E., Teggatz, P., Barman, M. & Salzman, N. H. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect. Immun. 77, 2741–2753 (2009).
Garner, C. D. et al Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection. Infect. Immun. 77, 2691–2702 (2009).
Ng, K. M. et al Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013).
Fernandez-Santoscoy, M. et al The gut microbiota reduces colonization of the mesenteric lymph nodes and IL-12-independent IFN-gamma production during Salmonella infection. Front. Cell. Infect. Microbiol. 5, 93 (2015).
Nagane, M. et al. Human glioblastoma xenografts overexpressing a tumor-specific mutant epidermal growth factor receptor sensitized to cisplatin by the AG1478 tyrosine kinase inhibitor. J. Neurosurg. 95, 472–479 (2001).
Ben-Arie, N. et al. Functional conservation of atonal and Math1 in the CNS and PNS. Development 127, 1039–1048 (2000).
Rose, M. F., Ahmad, K. A., Thaller, C. & Zoghbi, H. Y. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc. Natl Acad. Sci. USA 106, 22462–22467 (2009).
Shroyer, N. F. et al Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488 (2007).
Muller, A. J. et al The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6, 125–136 (2009).
Franchi, L. et al NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 13, 449–456 (2012).
Muzio, M., Ni, J., Feng, P. & Dixit, V. M. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 278, 1612–1615 (1997).
Cheminay, C., Mohlenbrink, A. & Hensel, M. Intracellular Salmonella inhibit antigen presentation by dendritic cells. J. Immunol. 174, 2892–2899 (2005).
van der Velden, A. W., Velasquez, M. & Starnbach, M. N. Salmonella rapidly kill dendritic cells via a caspase-1-dependent mechanism. J. Immunol. 171, 6742–6749 (2003).
Spadoni, I. et al A gut-vascular barrier controls the systemic dissemination of bacteria. Science 350, 830–834 (2015).
Chieppa, M., Rescigno, M., Huang, A. Y. & Germain, R. N. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med 203, 2841–2852 (2006).
Martinez-Argudo, I. & Jepson, M. A. Salmonella translocates across an in vitro M cell model independently of SPI-1 and SPI-2. Microbiology 154, 3887–3894 (2008).
Gautreaux, M. D., Deitch, E. A. & Berg, R. D. Bacterial translocation from the gastrointestinal tract to various segments of the mesenteric lymph node complex. Infect. Immun. 62, 2132–2134 (1994).
Lim, C. H. et al Independent bottlenecks characterize colonization of systemic compartments and gut lymphoid tissue by salmonella. PLoS Pathog. 10, e1004270 (2014).
Hapfelmeier, S. et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 174, 1675–1685 (2005).
Diehl, G. E. et al Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 494, 116–120 (2013).
Fierer, J., Okamoto, S., Banerjee, A. & Guiney, D. G. Diarrhea and colitis in mice require the Salmonella pathogenicity island 2-encoded secretion function but not SifA or Spv effectors. Infect. Immun. 80, 3360–3370 (2012).
Woo, H., Okamoto, S., Guiney, D., Gunn, J. S. & Fierer, J. A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PLoS ONE 3, e1603 (2008).
Ryan, C. A. et al. Massive outbreak of antimicrobial-resistant salmonellosis traced to pasteurized milk. JAMA 258, 3269–3274 (1987).
Pavia, A. T. et al. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J. Infect. Dis. 161, 255–260 (1990).
Dore, K. et al. Risk factors for Salmonella typhimurium DT104 and non-DT104 infection: a Canadian multi-provincial case-control study. Epidemiol. Infect. 132, 485–493 (2004).
Pabst, O. & Mowat, A. M. Oral tolerance to food protein. Mucosal Immunol. 5, 232–239 (2012).
Lathrop, S. K. et al Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011).
Nutsch, K. et al Rapid and efficient generation of regulatory T Cells to commensal antigens in the periphery. Cell Rep. 17, 206–220 (2016).
Franchi, L. et al Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7, 576–582 (2006).
Miao, E. A. et al Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107, 3076–3080 (2010).
Collazo, C. M. & Galan, J. E. The invasion-associated type-III protein secretion system in Salmonella--a review. Gene 192, 51–59 (1997).
Kaniga, K., Bossio, J. C. & Galan, J. E. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13, 555–568 (1994).
Galan, J. E., Lara-Tejero, M., Marlovits, T. C. & Wagner, S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438 (2014).
Kim, K. S. et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. https://doi.org/10.1126/science.aac5560 (2016).
Wlodarska, M. et al Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79, 1536–1545 (2011).
Glaccum, M. B. et al Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J. Immunol. 159, 3364–3371 (1997).
Hou, B., Reizis, B. & DeFranco, A. L. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity 29, 272–282 (2008).
Barnden, M. J., Allison, J., Heath, W. R. & Carbone, F. R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76, 34–40 (1998).
Kim, J. M., Rasmussen, J. P. & Rudensky, A. Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
el Marjou, F. et al Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Lee, T. C. & Threadgill, D. W. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis 47, 85–92 (2009).
McKinney, J., Guerrier-Takada, C., Galan, J. & Altman, S. Tightly regulated gene expression system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 6056–6059 (2002).
Hassan, J. O., Porter, S. B. & Curtiss, R. 3rd Effect of infective dose on humoral immune responses and colonization in chickens experimentally infected with Salmonella typhimurium. Avian Dis. 37, 19–26 (1993).
Vijay-Kumar, M. et al Flagellin suppresses epithelial apoptosis and limits disease during enteric infection. Am. J. Pathol. 169, 1686–1700 (2006).
Ben Hassena, A. et al. Real time PCR gene profiling and detection of Salmonella using a novel target: the siiA gene. J. Microbiol. Methods 109, 9–15 (2015).
Acknowledgements
Supported by grants: DK097317, DK109006, AI131342, AI077600, and Crohn’s and Colitis Foundation Research Fellowship Award 348359. The Washington University Digestive Diseases Research Center Core, supported by NIH grant P30 DK052574 assisted with imaging. The High Speed Cell Sorter Core at the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, provided flow cytometric cell sorting services. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant P30 CA91842.
Author information
Authors and Affiliations
Contributions
D.H.K., K.G.M., K.A.K., J.K.G., and K.M.K. performed the experiments. D.H.K., D.A.H., M.J.M., and R.D.N. designed the study. D.H.K, D.A.H., M.J.M., and R.D.N. wrote the manuscript. All authors have reviewed and agree with the manuscript content.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kulkarni, D.H., McDonald, K.G., Knoop, K.A. et al. Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol 11, 1103–1113 (2018). https://doi.org/10.1038/s41385-018-0007-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0007-6
This article is cited by
-
The role of goblet cells in Crohn’ s disease
Cell & Bioscience (2024)
-
The role of goblet cells and mucus in intestinal homeostasis
Nature Reviews Gastroenterology & Hepatology (2022)
-
Dendritic cell functions in the inductive and effector sites of intestinal immunity
Mucosal Immunology (2022)
-
Intestinal immune compartmentalization: implications of tissue specific determinants in health and disease
Mucosal Immunology (2021)
-
Murine astrovirus tropism for goblet cells and enterocytes facilitates an IFN-λ response in vivo and in enteroid cultures
Mucosal Immunology (2021)