Abstract
Acute myelogenous leukemia (AML) is the most common acute leukemia in adults. Despite great progress has been made in this field, the pathogenesis of AML is still not fully understood. We report here the biological role of lncRNA small nucleolar RNA host gene 5 (SNHG5) in the pathogenesis of AML and the underlying mechanisms. The results showed that lncRNA SNHG5 was highly expressed in AML cancer cell lines. In vitro studies displayed that inhibition of SNHG5 with shRNA resulted in suppression of survival, cell cycle progression, migration/invasion of AML and capacity of adhesion and angiogenesis in human umbilical vein endothelial cells. Mechanistic studies revealed a SNHG5/miR-26b/connective tissue growth factor (CTGF)/vascular endothelial growth factor A (VEGFA) axis in the regulation of AML angiogenesis. Finally, Yin Yang 1 (YY1) was found to transactivate and interact with SNHG5 promoter, leading to the upregulation of SNHG5 in AML. Collectively, upregulation of lncRNA SNHG5 mediated by YY1, activates CTGF/VEGFA via targeting miR-26b to regulate angiogenesis of AML. Our work provides new insights into the molecular mechanisms of AML.
Similar content being viewed by others
Introduction
Acute myelogenous leukemia (AML) is a cancer of the blood and bone marrow and is characterized by the arrest of differentiation and uncontrolled proliferation of myeloid blasts because of multiple epigenetic dysregulations and genetic mutations [1]. It is the most common acute leukemia among adults with 80% of adult patients with acute leukemia have AML, and the incidence of AML increases with age [2]. Although most of the patients achieve initial remission with the current chemotherapy, AML is still fatal in many younger patients and older adults because of therapy-related mortality, primary resistance, and relapse [3]. Considering these alarming indicators, a better understanding of the molecular mechanisms underlying the pathogenesis of AML is urgently needed.
Long noncoding RNAs (lncRNAs) are transcripts with size >200 nucleotides. LncRNAs may act in both cytoplasmic and nuclear compartments through different mechanisms [4]. LncRNAs are classified as intron lncRNAs, intergenic lncRNAs, antisense lncRNAs, and sense lncRNAs [5]. LncRNAs are involved in numerous biological roles including imprinting, epigenetic regulation, apoptosis, and cell cycle. Recently, lncRNAs have been reported to be dysregulated in AML and involved in the pathogenesis of AML [6]. Small nucleolar RNA host gene 5 (SNHG5) expression levels were upregulated in the plasma and bone marrow of patients with AML than that in the healthy cohorts. It is an independent prognostic biomarker as that the overall survival for those with high plasma SNHG5 expression was significantly shorter than those patients with low SNHG5 expression [7]. Yin Yang 1 (YY1) is a transcriptional factor with a dual function that regulate transcriptional activation and repression of multiple genes involved in many cellular processes including proliferation, differentiation, autophagy, and DNA repair [8]. Bioinformatics analysis predicted that YY1 potentially transactivates SNHG5 and miR-26b may directly interact with SNHG5. MiR-26b functions as a tumor suppressor in the development of T-cell acute lymphatic leukemia (T-ALL) as evidenced by the decreased level in mouse and human T-ALL cells and inhibition of T-ALL progression by miR-26b [9].
Connective tissue growth factor (CTGF) is a secreted matricellular protein, which has important roles in cancer cell migration, invasion, angiogenesis, and anoikis [10]. CTGF expression has been shown to be associated with the development and progression of many tumor types, such as poor prognosis in esophageal adenocarcinoma [11] and positive correlation with bone metastasis in breast cancer [12]. Recent studies have shown that CTGF expression is elevated in precursor-B-ALL. Moreover, increased expression of CTGF is associated with inferior outcomes in B-ALL [13]. Vascular endothelial growth factor (VEGF) is a homodimeric and heparin-binding glycoprotein. It is a critical pro-angiogenic protein in the physiological but also in most pathological angiogenesis, such as pathogenic processes of cancer [14]. It has been shown that CTGF increased the expression of VEGF by regulating miR-210 expression [15].
In this study, we identified that lncRNA SNHG5 was upregulated in AML cells and knockdown of SNHG5 inhibits angiogenesis of AML. Mechanistic studies revealed a YY1-SNHG5-miR-26b-CTGF-VEGFA axis in the regulation of AML angiogenesis.
Materials and methods
Cell lines and cell culture
Human AML cells U937, KG-1, THP-1, and HL-60 were purchased from ATCC (Manassas, VA, USA) and NB4 cells were obtained from Institutes for Biological Sciences (Shanghai, China). Cells were grown in Roswell Park Memorial Institute-1640 medium (RPMI-1640; Sigma-Aldrich) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA) and 1% penicillin and streptomycin (Sigma-Aldrich). Besides, CD34+ cells in bone marrow from four healthy volunteers (female/male = 1/1, 27–39 years old) were used as the normal control to these AML cell lines, and this experiment was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Nanchang University. All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
Plasmid constructions and cell transfection
The short hairpin RNA (shRNA) expression vectors targeting YY1 (sh-YY1: 5′-GCTCCAAGAACAATAGCTTGC-3′), VEGFA (sh-VEGFA: 5′-CGAACGTACTTGCAGATGTGA-3′) and SNHG5 (sh-SNHG5: 5′-CAAAGTTGCAACGATTTCTGG-3′), as well as YY1 overexpression vector (pcDNA3.1-YY1) were constructed as described previously [16,17,18]. MiRNA mimics (5′-UUCAAGUAAUUCAGGAUAGGU-3′) and inhibitors (5′-ACCUAUCCUGAAUUACUUGAA-3′) for miR-26b, negative control oligonucleotides (mimics NC: 5′-UUCUCCGAACGUGUCACGUTT-3′, inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′) were purchased from RiboBio (Guangzhou, China). For cell transfection, Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) was used in accordance with the manufacturer’s instructions. After 48 h of transfection, cells were performed for subsequent experiments. To construct the firefly luciferase reporter plasmids, the wild-type (WT) or mutant type (MUT) of the 3′-UTR regions of CTGF that was predicted to bind to miR-26b were synthesized from Sangon (Shanghai, China) and inserted into the downstream of the Renilla luciferase gene of pGL3 vectors (Promega, Madison, WI, USA).
Cell viability assay
The survival of cells was measured with the Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China) according to the manufacturer’s instructions. Cells were cultured at 3 × 103 cells/well in 96-well plates and CCK-8 assay was carried out after cell incubation for 24, 48, and 72 h. CCK-8 solution (10 μL) was added to cells and further incubated for 4 h at 37 °C in the dark. The optical density (OD) was assessed with a spectrophotometer (Thermo, USA) at 450 nm.
Colony formation assay
Cells were cultured at 1000 cells/well in a 6-well culture plate, and each experiment was performed in triplicate. The cells were grown for 14 days and colonies containing ≥50 cells were numbered under a microscope.
Flow cytometric analysis
Cell cycle analysis by quantitation of DNA content were performed with annexin V-fluorescein isothiocyanate and propidium iodine (PI) staining (Roche Diagnostics, Basel, Switzerland) according to the manufacturer’s instructions. The cells were collected and fixed with 70% cold ethanol for one hr at 4 °C. The cells were stained with PI of 1 mg/mL and were analyzed by flow cytometry (FACSCanto II, BD Biosciences). The data were analyzed using FlowJo software (LLC, Ashland, OR, USA).
Transwell assay
The cell migration and invasion activity were determined with the transwell chambers (BD Biosciences) with the membrane coated with or without Matrigel (Sigma). Briefly, cell suspension (serum free) was added into the upper transwell chamber, while the lower chamber was soaked with cell culture medium containing 10% FBS. After 12 h incubation, the chambers were fixed with 4% paraformaldehyde and stained with crystal violet subsequently. The cells which passed through the membrane were counted visually under a microscope (OLYMPUS).
Cell adhesion assay
After transfection, AML cells were cultured with serum-free medium for 48 h, and then the medium was collected as a conditioned medium. For cell adhesion assay, human umbilical vein endothelial cells (HUVECs) were seeded into each well in a 24-well plate and incubated with conditioned medium at 37 °C for 1, 2, and 4 h. The non-adherent cells were washed twice with phosphate-buffered saline (PBS) and the attached cells were measured by an optical microscope.
Endothelial tube formation assay
Cell culture medium from AML cells was collected as described above, and HUVECs were cultured with the cell culture medium for 24 h. HUVECs were cultured at a concentration of 1 × 104 cells/well in a 96-well plate, which was precoated with Matrigel (50 μL/well) for 5 h. The tubules analyzed by ImageJ software.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The cDNA was obtained using a first-strand cDNA kit (Sigma, Munich, Germany), according to the manufacturer’s protocol. PCR amplification was performed using an SYBR Green PCR kit (Thermo) and run on an ABI 7300 Thermocycler (Applied Biosystems, Foster City, CA, USA). The U6 small nuclear RNA and GAPDH was amplified as a loading control, respectively. The relative expression levels of genes were assessed by using the 2−∆∆Ct method. The sequences of specific primers used in this study were as follows:
SNHG5: 5′-CACAGTGGAGCAGCTCTGAA-3′ (forward), SNHG5: 5′-CTCGTGGCACTAGCCAGAAA-3′ (reverse); MIR-26B: 5′-UCAAGUAAUUCAGGAUAGGU-3′ (forward), MIR-26B: 5′-CGGCCCAGTGTTCAGACTAC-3′ (reverse); CTGF: 5′-CGACTGGAAGACACGTTTGG-3′ (forward), CTGF: 5′-TGGGAGTACGGATGCACTTT-3′ (reverse); YY1: 5′-AAAACATCTGCACACCCACG-3′ (forward), YY1: 5′-GTCTCCGGTATGGATTCGCA-3′ (reverse); U6: 5′-CTCGCTTCGGCAGCACA-3′ (forward), U6: 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); GAPDH: 5′-CCAGGTGGTCTCCTCTGA-3′ (forward), GAPDH: 5′-GCTGTAGCCAAATCGTTGT-3′ (reverse).
Western blot analysis
Total proteins were extracted using RIPA Buffer (Cell Signaling Technology, Danvers, MA, USA). Proteins were separated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (EMD Millipore). The membranes were subsequently blocked with 5% nonfat milk for 2 h at room temperature and immunoblotted overnight at 4 °C with the primary antibodies. Then the membranes were incubated with secondary antibody for 2 h at room temperature. Anti-CD31 (cat. no. ab134168; dilution, 1000), anti-VEGFA (cat. no. ab46154; dilution, 1000), anti-CTGF (cat. no. ab231824; dilution, 1000), and anti-β-actin (cat. no. ab8227; dilution, 1:5000) antibodies were from Abcam. Secondary horseradish peroxidase-conjugated goat anti-rabbit (dilution, 1:10,000; cat. no. ab205718) antibodies were used. The bands were developed with RapidStep™ ECL Reagent (EMD Millipore) according to the manufacturer’s directions.
RNA immunoprecipitation (RIP) assay
RIP assay was performed according to the manufacturer’s protocol (Millipore Corporation). In brief, cells were harvested and lysed followed by incubation with anti-Ago2 or anti-IgG at 4 °C overnight with gentle rotation. The protein A/G beads (40 µL) were added and incubated with lysate for 1 hr at 4 °C to capture the complex. After washing in the buffer, RNA was isolated for detection of its concentration by qRT‐PCR.
Dual-luciferase reporter assay
The reporters were transfected into cells and the luciferase activity was assessed using a Dual-Glo® Luciferase kit according to the manufacturer’s instructions. The activities of firefly luciferase and Renilla luciferase were measured, with the latter as the internal control.
Chromatin immunoprecipitation (ChIP) assay
ChIP analysis was performed with ChromaFlash High-Sensitivity ChIP Kit (Epigentek, #P-2027-24). In brief, cells were treated with 1.0% formaldehyde to covalently link DNA and protein. After cross-linked, cells are lysed in 0.1% SDS containing buffer to obtain DNA fragmentation. DNA-protein samples were incubated with the anti-YY1 antibody (ab232573, Abcam) overnight at 4 °C by constant rotation in the cold room followed by manufacturer’s protocol. Co-precipitated DNAs were analyzed by qRT-PCR.
Immunohistochemistry (IHC)
To examine the expressions of CD31 and Ki-67 in tumor tissues, IHC assay was carried out. In brief, sections were deparaffinized, rehydrated, and blocked with 10% normal rabbit serum in PBS. The tissue sections were incubated sequentially with a CD31 primary antibody (cat. no. ab134168, Abcam, USA) or a Ki-67 primary antibody (cat. no. ab15580, Abcam, USA) for 30 min, followed by incubation with biotin-conjugated goat anti-rabbit IgG (Solarbio, Shanghai) for 30 min, horseradish peroxidase–streptavidin (Solarbio, Shanghai) for additional 30 min and the immunoreactivity was developed using diaminobenzidine reagent (Vector Laboratories, Burlingame, USA) for 10 min. Images were obtained with an Leica DM6000B microscope (Leica Microsystems, Germany).
Tumor xenografts
Transfected NB4 cells were subcutaneously implanted into the right flank of 5-week old nude mice (10 mice, 5 in each group). When the tumor volume was ~400–600 mm [3], the tumors were collected for subsequent analysis including qRT-PCR, western blot, and IHC. Experimental procedures involving animals were performed in accordance with “Principles of Laboratory Animal Care” (NIH publication no. 85–23, revised 1985).
Statistical analysis
All data were presented as the mean of at least triplicate samples ± standard deviation (SD). Statistical analysis was performed with the SPSS statistical software program (version 13.0; SPSS, Inc., Chicago, IL, USA). Differences between groups were analyzed by the Student’s t test or one-way analysis of variance. P < 0.05 was considered statistically significant.
Results
The expression of lncRNA SNHG5 and miR-26b in AML cell lines
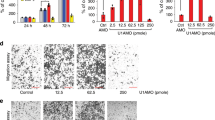
First, expression of lncRNA SNHG5 and miR-26b was analyzed by qRT-PCR in AML cell lines and the normal control. The results revealed that the expression level of SNHG5 was higher in all five AML cells (U937, KG-1, NB4, THP-1, and HL-60) when compared with that of control cells (Fig. 1a). In contrast, the level of miR-26b was decreased in AML cells than that of normal control (Fig. 1b).
a The expression level of SNHG5 was detected in AML cell lines and bone marrow CD34+ cells from health donors by using qRT-PCR. b The expression of miR-26b was detected in AML cell lines and normal control cells by using qRT-PCR. “**” denotes p < 0.01; “***” denotes p < 0.001. Data are expressed as the mean ± SD.
LncRNA SNHG5 affects AML-associated angiogenesis
To explore the potential biological roles of SNHG5 in AML, the effects of SNHG5 silencing on AML cells (THP-1 and NB4) were investigated. The cell lines transfected with sh-SNHG5 showed a significant reduction in endogenous SNHG5 expression (Fig. 2a). Cell survival curves analyzed by CCK-8 assay demonstrated time-dependent suppression of cell viability upon knockdown of SNHG5 (Fig. 2b). Consistently, colony formation assay confirmed this data by validated that SNHG5 inhibition resulted in a reduction in the clonogenic survival of AML cells relative to the control group (Fig. 2c). Knockdown of SNHG5 caused the arrest of the cell cycle as shown by a decrease in the proportion of cells in the S phase and the dramatic increase in the proportion of cells in G1 (Fig. 2d). Furthermore, cell migration and invasion activity were remarkably inhibited when SNHG5 was downregulated (Fig. 2e). As shown in Fig. 2f, the results of western blot analysis showed that knockdown of SNHG2 resulted in the downregulation of the protein level of CD31 and VEGFA. In addition, downregulation of SNHG5 induced time-dependent inhibition of adhesion of HUVECs as measured by cell adhesion assay (Fig. 2g). Silencing of SNHG5 further caused a decrease in tube formation of HUVECs (Fig. 2h). Taken together, these results suggested that SNHG5 promotes cell survival, cell cycle progression, cell migration/invasion, and cell adhesion as well as angiogenesis.
a The expression of SNHG5 was detected in both AML cell lines by using RT-qPCR. Effects of SNHG5 silencing on cell viability (b), cell proliferation (c), cell cycle (d), cell migration, and invasion (e) in AML cells were measured by CCK-8, colony formation, FACS, and Transwell assays, respectively. f The protein expression of CD31 and VEGFA was detected using western blot analysis. g Effect of SNHG5 silencing on cell adhesion was measured by cell adhesion assay. h The representative image of the tube formation of HUVECs was assessed with endothelial tube formation assay. “*” denotes p < 0.05. “**” denotes p < 0.01. Data are expressed as the mean ± SD.
LncRNA SNHG5 functions as a sponge for miR-26b
Several lncRNAs were reported to serve as competing endogenous RNAs (ceRNAs) by competitively binding miRNAs. To further examine the potential mechanism underlying the function of SNHG5 in AML, we speculated that miRNAs participate in the biological process of AML. Bioinformatic prediction with starBase software predicted that SNHG5 has a high probability of binding sites for miR-26b (Fig. 3c). Knockdown of SNHG5 resulted in an increase of miR-26b in AML cells (Fig. 3a). The miR-26b mimics and inhibitors were further used to change the expression of miR-26b in cells (Fig. 3b). The results of RIP assay showed that overexpression of miR-26b enriched the binding of SNHG5 to Ago2-containing micro-ribonucleoprotein complexes, which confirmed that SNHG5 was able to bind to miR-26b in both AML cells (Fig. 3d). To further verify SNHG5 was physically associated with miR-26b, SNHG5 3′-UTR containing the miR-26b-binding sites were cloned downstream of the luciferase open-reading frame to obtain constructs for luciferase activity assay. The results showed that overexpression of miR-26b significantly downregulated luciferase activity, however, mutant abolished the above effects (Fig. 3e). These results indicated that SNHG5 functions as a sponge for miR-26b.
a The expression of miR-26b was detected in sh-SNHG5 transfected cells and control cells by using RT-qPCR. b The expression of miR-26b was detected in AML cell lines transfected with miR-26 mimics, miR-26b inhibitor by using qRT-PCR. c Bioinformatics analysis showed the prediction for miR-26b bindings sites on the 3′-UTR of SNHG5. d RIP assay confirmed that lncRNA SNHG5 could bind to miR-26b. e Luciferase activity in AML cells overexpressing of miR-26b and co-transfection of luciferase reporters containing SNHG5 3′-UTR or mutant. “*” denotes p < 0.05. “**” denotes p < 0.01; “***” denotes p < 0.001. Data are expressed as the mean ± SD.
LncRNA SNHG5 acts as a ceRNA and regulates CTGF expression by competitively binding miR-26b
As it has been predicted that CTGF was a target of miR-26b by starBase software, RT-qPCR was performed to determine the expression level of CTGF in AML cells. The results showed that CTGF was upregulated in AML cells when compared with normal control (Fig. 4a). As displayed in Fig. 4b, overexpression of miR-26b markedly suppressed CTGF expression in AML cells. The 3′-UTR of CTGF with miR-26b-binding sites were cloned downstream of the luciferase open-reading frame to obtain constructs for luciferase activity assay (Fig. 4c). The results showed that overexpression of miR-26b significantly downregulated luciferase activity. However, the mutant abolished the above effects (Fig. 4d). To further verify that whether SNHG5 regulated the expression of CTGF by targeting miR-26b, sh-SNHG5 treated cells were transfected with miR-26b inhibitor or negative control. The data showed that downregulation of SNHG5 resulted in decrease of CTGF, however, inhibition of miR-26b showed partial abrogation of the above effects (Fig. 4e). These results indicated that SNHG5 knockdown suppresses the downstream targets of CTGF via sponging miR-26b.
a The expression of CTGF was detected in AML cells using qRT-PCR and western blot. b The mRNA and protein levels of CTGF were detected in AML cells transfected with miR-26b or negative control. c Bioinformatics analysis showed the prediction for miR-26b bindings sites on the 3′-UTR of CTGF. d Luciferase activity in AML cells overexpressing of miR-26b and co-transfection of luciferase reporters containing SNHG5 3′-UTR or mutant. e The expression of CTGF in AML cells upon treatment of sh-SNHG5 with miR-26b inhibitor or negative control was detected using qRT-PCR and western blot, respectively. “**” denotes p < 0.01; “***” denotes p < 0.001. Data are expressed as the mean ± SD.
LncRNA SNHG5 regulates AML-associated angiogenesis through miR-26b/CTGF axis
Since it has been shown that CTGF increased expression of VEGFA by regulating miR-210 expression [15], we speculated that SNHG5 regulates angiogenesis of AML by regulating miR-26b/CTGF/VEGFA axis. The data of colony formation assay showed that miR-26b inhibition resulted in the promotion of cell proliferation, whereas knockdown of SNHG5 or VEGFA abolished the effect of miR-26b suppression on cell proliferation (Fig. 5a). MiR-26b inhibitor treatment caused increased cell number in S phase and the dramatic decrease in the proportion of cells in G0/G1 phase in AML cells. In contrast, knockdown of SNHG5 or VEGFA abolished the effect of miR-26b decrease on cell cycle progression (Fig. 5b). Besides, miR-26b inhibitor treatment resulted in the promotion of cell migration and invasion ability. However, knockdown of SNHG5 or VEGFA reversed these changes (Fig. 5c, d). Moreover, HUVECs angiogenesis was found to be increased upon treatment of miR-26b inhibitor, whereas knockdown of SNHG5 or VEGFA abolished the AML-associated angiogenesis (Fig. 5e). In summary, these data indicated that SNHG5 regulates angiogenesis via miR-26b/CTGF axis.
The cells were treated with miR-26b inhibitor and sh-SNHG5 or sh-VEGFA followed by the detection of cell proliferation a with colony formation assay, cell cycle analysis (b) with FACS, assessment of cell migration (c), and invasion (d) using Transwell assay, and measurement of angiogenesis (e) with endothelial tube formation assay. “*” denotes p < 0.05; “**” denotes p < 0.01; “***” denotes p < 0.001. Data are expressed as the mean ± SD.
YY1 activates the transcription of lncRNA SNHG5
To identify upstream regulators that contributed to the overexpression of SNHG5 in AML, bioinformatics analysis predicted whether YY1 potentially transactivates SNHG5 by JASPAR database (http://jaspar.genereg.net/). Upregulation of YY1 in AML cells was observed when compared with normal cells (Fig. 6a). Knockdown YY1 induced the downregulation of SNHG5 in both of AML cells. Conversely, overexpression of YY1 resulted in upregulation of SNHG5 (Fig. 6b). As shown in Fig. 6c, d, the two potential putative binding sites upstream of SNHG5 named P1 and P2 were predicted. ChIP assay was carried out by using the specific YY1 antibody, showing that YY1 directly bound to the presumed P2 sites instead of P1 (Fig. 6e). In addition, dual-luciferase reporter assay was performed by co-transfecting PGL3 reporter containing the P2-binding sites of YY1 alone or together with pcDNA3.1-YY1 plasmid into AML cells. The results indicated that YY1 promoted the transcriptional activity of SNHG5 (Fig. 6f). In summary, YY1 binds to the promoter of SNHG5 and activates SNHG5 transcription.
a YY1 expression in AML cells assessed with qRT-PCR analysis. b The expression of YY1 and SNHG5 in cells transfected with sh-YY1 or shRNA control as well as pcDNA3.1-YY1 or pcDNA3.1-NC. c The binding motif of transcription factor YY1 from JASPAR database. d The two potential putative binding sites upstream of SNHG5 named P1 and P2 were predicted. e The enrichment of YY1 on the promoter of SNHG5 was assessed with ChIP assay. f Luciferase activity in AML cells overexpressing of YY1 and co-transfection of luciferase reporters containing SNHG5. “*” denotes p < 0.05; “**” denotes p < 0.01; “***” denotes p < 0.001. Data are expressed as the mean ± SD.
LncRNA SNHG5 regulates angiogenesis in a mouse model of AML
The sh-SNHG5 transfected NB4 cells were subcutaneously injected into the nude mice and the tumors were collected and analyzed. The expression level of miR-26b was upregulated and CTGF mRNA expression level was downregulated when SNHG5 expression was inhibited (Fig. 7a). In addition, the protein level of VEGFA, CD31 and Ki-67 was lower in sh-SNHG5 cells when compared with control group (Fig. 7b). In line with the western blot data, the result of IHC analysis showed that inhibition of SNHG5 suppressed the staining of Ki-67 and CD31 as well as the microvessel density value (Fig. 7c). These data indicated that SNHG5 regulates angiogenesis in vivo.
Transfected NB4 cells were subcutaneously injected into the nude mice and the tumors were collected and analyzed. a The expression levels of miR-26b, CTGF, and SNHG5 in tumor tissues of mice. b Western blotting analyses of CD31, Ki-67, and VEGFA in tumor tissues. c Representative image of IHC staining of Ki-67 and CD31 and the quantification of CD31 and Ki-67. “*” denotes p < 0.05; “**” denotes p < 0.01; “***” denotes p < 0.001. Data are expressed as the mean ± SD.
Discussion
AML is the most common acute leukemia in adults. Despite great progress has been made in this field, the pathogenesis of AML is still not fully understood [19]. In this study, we identify that lncRNA SNHG5 is activated by YY1 in AML cells and knockdown of SNHG5 inhibits angiogenesis of AML via SNHG5-miR-26b-CTGF-VEGFA axis.
LncRNAs are transcripts >200 nt, which play a key role in tumorigenesis and cancer progression of AML. For example, LINP1 was significantly overexpressed in AML patients at diagnosis and promoted the malignant phenotype of AML cells [20]. LncRNA-H19 was found to be upregulated in AML and promote the proliferation of AML cells, but inhibit the apoptosis [21]. SNHG5 was located on chromosome 6q15 at the chromosomal translocation breakpoint. It was functionally implicated in multiple tumor types and is correlated with tumor progression, including gastric cancer [22], chronic myeloid leukemia [23], melanoma [24], colorectal cancer [25], breast cancer [26], hepatocellular carcinoma [16], and bladder cancer [27]. Nevertheless, the roles and molecular bases of SNHG5 in pathogenesis of AML are not well understood. In the process of tumorigenesis and development, there is a crucial step, called tumor angiogenesis. Tumor cells also often invade these new blood vessels and metastasize to other organs through the blood. Therefore, angiogenesis is closely related to the degree of malignancy. In this study, we for the first time reported that SNHG5 was involved in the cell proliferation, cell cycle progression, cell migration and invasion, cell adhesion, and angiogenesis of AML. These findings were consistent with above previous studies in many tumors.
Furthermore, there was a ceRNA hypothesis that proposed that lncRNAs may compete for binding to miRNAs so as to regulate target gene expression and play an important role in various diseases [26]. SNHG5 was considered as a ceRNA of miR-26a [24], miR-154-5p [26], miR-212-3p [28], and miR-205-5p [23]. In this research, with the aim of identifying miRNAs that are regulated by SNHG5, we found miR-26b based on bioinformative analysis. With various assays, the direct interaction between SNHG5 and miR-26b was further confirmed. Decreased expression of miR-26b was found in many cancers such as locally advanced breast cancer [29], colorectal cancer [30], and cervical cancer [31] and was found to be a tumor suppressor. Consistently, in this study miR-26b was decreased in AML cells and inhibition of miR-26b contributed to tumor progression of AML, including proliferation, migration, invasion, and angiogenesis. CTGF expression has been shown to be associated with the development and progression of many tumor types [10]. It has been shown that CTGF increased the expression of VEGFA by regulating miR-210 expression [15]. In this study, we reported that miR-26b directly targets CTGF and regulates VEGFA via targeting CTGF. VEGFA is well known as a critical regulator for tumor angiogenesis, which is highly expressed in many tumors and its expression level is related to tumor stage and progression. In the present study, we also found that VEGFA suppression could partially reverse the promotion effects of miR-26b inhibition on cell proliferation, migration, invasion, and angiogenesis, suggesting VEGFA may be involved in AML pathogenesis. Collectively, lncRNA SNHG5 may regulate AML-associated angiogenesis through CTGF/VEGFA axis by targeting miR-26b.
YY1 is a transcriptional factor that is an important factor of GLI-Kruppel class of zinc finger proteins [8]. YY1 is increased in many malignant diseases. Moreover, YY1 is able to modulate the expression of both protein-coding genes and noncoding genes in human cancer cells [32]. Consistently, we found that YY1 was upregulated in AML cells. Bioinformatics analysis predicted that YY1 potentially transactivates SNHG5. The data of ChIP assay and luciferase reporter assay demonstrated that YY1 transactivate and bind to the SNHG5 promoter, which at least partially explained the overexpression of SNHG5 in AML cells.
This study is the first to illustrate that upregulation of SNHG5 mediated by YY1, activates CTGF by targeting miR-26b to regulate angiogenesis of AML. In conclusion, our work provides new insights into the molecular mechanisms of AML. Based on these evidence, therapeutic strategies would be directed toward the downregulation of SNHG5, which may potentially be a clinically viable target in the treatment of AML.
References
Bi L, Zhou B, Li H, He L, Wang C, Wang Z, et al. A novel miR-375-HOXB3-CDCA3/DNMT3B regulatory circuitry contributes to leukemogenesis in acute myeloid leukemia. BMC Cancer. 2018;18:182.
Short NJ, Rytting ME, Cortes JE. Acute myeloid leukaemia. Lancet. 2018;392:593–606.
Liu Y, Lei P, Qiao H, Sun K, Lu X, Bao F, et al. miR-9 enhances the chemosensitivity of AML cells to daunorubicin by targeting the EIF5A2/MCL-1 axis. Int J Biol Sci. 2019;15:579–86.
Morlando M, Ballarino M, Fatica A. Long non-coding RNAs: new players in hematopoiesis and leukemia. Front Med (Lausanne). 2015;2:23.
Liu Y, Cheng Z, Pang Y, Cui L, Qian T, Quan L, et al. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J Hematol Oncol. 2019;12:51.
Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci USA. 2014;111:18679–84.
Li J, Sun CK. Long noncoding RNA SNHG5 is up-regulated and serves as a potential prognostic biomarker in acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2018;22:3342–7.
Sarvagalla S, Kolapalli SP, Vallabhapurapu S. The two sides of YY1 in cancer: a friend and a foe. Front Oncol. 2019;9:1230.
Yuan T, Yang Y, Chen J, Li W, Li W, Zhang Q, et al. Regulation of PI3K signaling in T-cell acute lymphoblastic leukemia: a novel PTEN/Ikaros/miR-26b mechanism reveals a critical targetable role for PIK3CD. Leukemia. 2017;31:2355–64.
Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci. 2008;15:675–85.
Ustiuzhaninova NV, Shishkin GS. [Dimensions and alveolar surface of the ineralveolar septa of the rat lung]. Arkh Anat Gistol Embriol. 1975;68:59–63.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cardo CC, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49.
Lu H, Kojima K, Battula VL, Korchin B, Shi Y, Chen Y, et al. Targeting connective tissue growth factor (CTGF) in acute lymphoblastic leukemia preclinical models: anti-CTGF monoclonal antibody attenuates leukemia growth. Ann Hematol. 2014;93:485–92.
Melincovici CS, Boşca AB, Şuşman S, Mărginean M, Mihu C, Istrate M, et al. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol. 2018;59:455–67.
Liu SC, Chuang SM, Hsu CJ, Tsai CH, Wang SW, Tang CH. CTGF increases vascular endothelial growth factor-dependent angiogenesis in human synovial fibroblasts by increasing miR-210 expression. Cell Death Dis. 2014;5:e1485.
Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y, et al. Long non-coding RNA SNHG5 promotes human hepatocellular carcinoma progression by regulating miR-26a-5p/GSK3beta signal pathway. Cell Death Dis. 2018;9:888.
Wu S, Wang H, Li Y, Xie Y, Huang C, Zhao H, et al. Transcription factor YY promotes cell proliferation by directly activating the pentose phosphate pathway. Cancer Res. 2018;78:4549–62.
Qiu JF, Zhang ZQ, Wang Y, You J. Lentivirus-mediated RNAi knockdown of VEGFA in RKO colorectal cancer cells decreases tumor formation and growth in vitro and in vivo. Int J Clin Exp Pathol. 2012;5:290–8.
Zhao TF, Jia HZ, Zhang ZZ, Zhao XS, Zou YF, Zhang W, et al. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol Med Rep. 2017;16:3687–93.
Shi J, Dai R, Chen Y, Guo H, Han Y, Zhang Y, et al. LncRNA LINP1 regulates acute myeloid leukemia progression via HNF4alpha/AMPK/WNT5A signaling pathway. Hematol Oncol. 2019;37:474–82.
Zhao TT, Liu X. LncRNA-H19 inhibits apoptosis of acute myeloid leukemia cells via targeting miR-29a-3p. Eur Rev Med Pharmacol Sci. 2019;23:224–31.
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31:893–903.
He B, Bai Y, Kang W, Zhang X, Jiang X. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res. 2017;7:1704–13.
Gao J, Zeng K, Liu Y, Gao L, Liu L. LncRNA SNHG5 promotes growth and invasion in melanoma by regulating the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. 2019;12:169–79.
Damas ND, Marcatti M, Côme C, Christensen LL, Nielsen MM, Baumgartner R, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875.
Chi JR, Yu ZH, Liu BW, Zhang D, Ge J, Yu Y, et al. SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA Axis. Mol Ther Nucleic Acids. 2019;17:138–49.
Ma Z, Xue S, Zeng B, Qiu D. lncRNA SNHG5 is associated with poor prognosis of bladder cancer and promotes bladder cancer cell proliferation through targeting p27. Oncol Lett. 2018;15:1924–30.
Ju C, Zhou R, Sun J, Zhang F, Tang X, Kwok K, et al. LncRNA SNHG5 promotes the progression of osteosarcoma by sponging the miR-212-3p/SGK3 axis. Cancer Cell Int. 2018;18:141.
Ding Q, Wang Y, Zuo Z, Gong Y, Krishnamurthy S, Li CW, et al. Decreased expression of microRNA-26b in locally advanced and inflammatory breast cancer. Hum Pathol. 2018;77:121–9.
Wang B, Lu FY, Shi RH, Feng YD, Zhao XD, Lu ZP, et al. MiR-26b regulates 5-FU-resistance in human colorectal cancer via down-regulation of Pgp. Am J Cancer Res. 2018;8:2518–27.
Wang L, Wang W, Wu Y. MicroRNA-26b acts as an antioncogene and prognostic factor in cervical cancer. Oncol Lett. 2019;17:3418–24.
Xia Y, Wei K, Yang FM, Hu LQ, Pan CF, Pan XL, et al. miR-1260b, mediated by YY1, activates KIT signaling by targeting SOCS6 to regulate cell proliferation and apoptosis in NSCLC. Cell Death Dis. 2019;10:112.
Acknowledgements
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Funding
This work was supported by the National Natural Science Foundation of China (no. 81860036).
Author information
Authors and Affiliations
Contributions
Guarantor of integrity of the entire study: Zhen-Jiang Li, Ji-Fu Zheng. Study concepts: Zhen-Jiang Li, Ji-Fu Zheng. Study design: Zhen-Jiang Li, Ji-Fu Zheng. Definition of intellectual content: Zhen-Jiang Li. Literature research: Zhen-Jiang Li, Jing Cheng. Experimental studies: Yuan Song. data acquisition: Jing Cheng. Data analysis: Zhen-Jiang Li. Statistical analysis: Hui-Hui Li. Manuscript preparation: Zhen-Jiang Li. Manuscript editing: Ji-Fu Zheng. Manuscript review: Ji-Fu Zheng. All authors approved the final version of the manuscript to be published.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, ZJ., Cheng, J., Song, Y. et al. LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. Lab Invest 101, 341–352 (2021). https://doi.org/10.1038/s41374-020-00519-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-00519-9
This article is cited by
-
Single-cell transcriptional landscape of long non-coding RNAs orchestrating mouse heart development
Cell Death & Disease (2023)
-
Long non-coding RNA SNHG5 promotes the osteogenic differentiation of bone marrow mesenchymal stem cells via the miR-212-3p/GDF5/SMAD pathway
Stem Cell Research & Therapy (2022)
-
HDAC1/3-dependent moderate liquid–liquid phase separation of YY1 promotes METTL3 expression and AML cell proliferation
Cell Death & Disease (2022)