Abstract
β-S globin haplotype (βS haplotype) characterization in sickle cell anemia (SCA) patients is important because it assists individualized treatment. However, the patient with atypical haplotypes do not present detailed studies such as clinical and laboratory data. To understand the phenotypic expression of atypical haplotype patients in relation to typical haplotype ones, it may be necessary to assess the main clinical and laboratorial parameters and investigate transcription factors, as possible genetic modulators that can contribute to the improvement of the SCA patients’ clinical condition. The study group was composed of 600 SCA Brazilian patients of both genders ranging in age from 1 to 68 years. The atypical haplotypes were the third most frequent (5.7%) with 11 patterns numerically ranked according to occurrence. We verified that patients with atypical 1 haplotype in combination with Bantu haplotype presented milder clinical outcomes in relation to Bantu/Bantu and Benin/Benin patients, according to improved values of hemoglobin and hematocrit. In clinical severity, we did not observe significant statistical differences between typical and atypical haplotype patients, and this result can be explained with reference to the action of transcription factors in β-globin cluster. Thus, we presented the atypical haplotype relationship with SCA pathophysiology, reinforcing the hypothesis that individual genetic factors may be responsible for phenotypic diversity of the disease.
Similar content being viewed by others
Introduction
Sickle cell disease (SCD) is the most common genetic complication in the world, and it is characterized by the hemoglobin S (Hb S) presence [1]. The Hb S mutation (HBB:c.20A>T −rs334) [2] probably emerged in the continent of Africa, where malaria was the main modulator. According to historical and genetic characteristics, researchers began to track polymorphic sites in order to study the anthropology of Hb S in Africa [3], and its origin and dissemination in other regions of the world [4]. Preliminary investigations contributed to the discovery of many polymorphic sites along the β-globin cluster [5], named β-S globin haplotypes (βS haplotypes). Therefore, the βS-haplotypes are a set of mutations in the β-globin cluster recognized by specific restriction enzymes, which are inherited together due to their proximity [6].
Currently, there are five typical βS-haplotypes described and they are designated according to the geographical area where they are most commonly found, being as follows: Benin associated with Midwestern Africa, Senegal with West Africa, Bantu with South-Central and Eastern Africa [5], the Arab-Indian-Saudita (Saudi) associated with the Indian subcontinent and the Eastern Arabian Peninsula [7], and Cameroon along the west coast of Africa [8]. During the βS-haplotype investigations, researchers found unusual haplotypes in relation to the patterns described, and they were named as atypical haplotypes [5, 9, 10]. The different haplotype patterns are generated by several genetic mechanisms involving βS-chromosomes containing typical haplotypes, such as point mutations and recombination or gene conversion [5, 9, 10].
Besides the importance of studies about population genetics, anthropology, origins, and evolution of Hb S mutation, additional investigations about the βS-haplotypes may be helpful to explain the great clinical diversity observed in sickle cell anemia (SCA) patients, giving direction to suitable individualized treatment [11, 12]. In the present study, we characterized the atypical haplotypes found in our SCA population. Furthermore, we compared the Hb F levels, the main hematologic and hemolytic markers and the clinical severity score among the SCA patients with at least one atypical haplotype in relation to the reference groups: Bantu/Bantu and Benin/Benin. Finally, we used in silico analysis to evaluate six polymorphic sites that characterize the βS-haplotype in order to verify the possible transcription factors (TFs) that can interfere with β-globin cluster modulation.
Materials and methods
Patients
The study group was selected from 1348 SCD patients regularly treated at the State Institute of Hematology “Arthur de Siqueira Cavalvanti”—HEMORIO. After the genotyping of hemoglobin profiles and βS-haplotypes, we included patients in the study with SCA (Hb SS) without any degree of kinship, and whose haplotype identification was conclusive. Therefore, the study group was composed of 600 SCA patients, 338 females and 262 males, with ages varying from 1 to 68 years old (median average of 22 years old). All patients consented to participate in this study which was approved by the Supervision and Data Security Board of the institution in accordance with the Brazilian Norms and Research Ethics Committee.
Blood samples
Peripheral blood samples were collected by means of venipuncture in two 4 mL tubes, each one containing 5% ethylenediaminetetraacetic acid. One tube was used to measure the hematological and hemolytic index. The other tube was used to characterize the hemoglobin profile and βS-haplotypes.
Sickle cell disease genotyping and βS haplotype characterization
Genomic DNA was extracted from leukocytes for molecular analysis [13]. The Hb S mutation (HBB:c.20A>T −rs334) was confirmed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and the amplified fragment was cleaved using DdeI (C↓TNAG) restriction enzyme (Thermo Fisher Scientific, Waltham, MA USA) [14]. The characterization of βS-haplotypes was performed by PCR-RFLP and the following polymorphic sites were analyzed: 5′-HBG2 (XmnI) HBG2 (HindIII), HBG1 (HindIII), HBBP (HincII), 3′-HBBP (HincII), and 5′-HBB (HinfI), as previously described [15].
Hb F levels, hematologic, and hemolytic index
Quantification of Hb F levels were obtained by high-performance liquid chromatography using automated equipment (Ultra [2], Trinity Biotech HPLC, Bray, Co Wicklow, Ireland). Hematologic markers were estimated by flow cytometry and spectrophotometry methods (Cell-Dyn Ruby, Abbott Laboratories, Chicago, IL, USA). The parameters measured were as follows: red blood cell count (RBC, M/µL), Hb levels (g/dL), hematocrit (Ht, %), mean corpuscular volume (MCV, fL), and relative reticulocyte (Rel. Ret., %). Serum total bilirubin (TB, mg/dL) levels were estimated by 2,4-dichlorophenyl-diazonium method. Lactate dehydrogenase (LDH, U/L) and aspartate aminotransferase (AST, U/L) were measured by ultraviolet enzymatic methods (AU680 Chemistry System, Beckman Coulter Inc., Jersey, NJ, USA). The Rel. Ret. was analyzed along with LDH, AST, and TB to infer hemolysis [16].
Clinical severity classification
We used the online tool “Sickle Cell Disease Severity Calculator,” available at http://www.bu.edu/sicklecell/downloads/Projects, to classify the SCA patients into categories according to clinical severity (mild, intermediate, and severe). The network model calculates the death risk within 5 years and classifies this risk on a disease severity score, ranging from 0 (least severe) to 1 (most severe). The calculator takes into account the following variables that are sufficient in order to compute the score for the severity of the disease: age, gender, TB, blood transfusion, reticulocyte count, LDH, MCV, total leukocyte, acute chest syndrome, pain crises, priapism, stroke, avascular necrosis of bone, and SCD genotype. Scores ranging from 0 to 0.399 were considered mild, from 0.400 to 0.599 intermediate, and from 0.600 to 1.0 severe [17]. This tool was standardized and validated by our research group for SCD Brazilian patients [18].
In silico TF detection
The dataset evaluated in this study comprised of the evaluation of approximately 200 bp sequences of three regions from the human HBB gene (NG_000007.3; one in HBG2, ID3048; two in HBBP, ID3044). The set of vertebrate TFs with their binding sites was obtained from the non-redundant, curated, and publicly available JASPAR database (http://jaspar.genereg.net) [19, 20]. The TF binding site (TFBS) analysis was performed in the XmnI (5′-HBG2), HincII (HBBP), and HincII (3′-HBBP) sites in order to identify TFs associated with these regions. In order to identify possible changes to TFBS due to the presence of a polymorphism, the evaluation was performed in two different situations: a TFBS search in sequences containing the ancestral allele (wild type) and a TFBS search in sequences containing the polymorphisms.
Statistical analysis
Statistical analysis was performed using the Statistica 8.0 software (StatSoft Inc., Tulsa, OK, USA) and the graphics were built using GraphPad Prisma version 5.01 for Windows (GraphPad Software, La Jolla, CA, USA). The data were tested for normality by using normal probability plots of residuals. To perform the statistical test, we removed the extreme values of the Ht, LDH, AST, TB, and Rel. Ret. parameters that were selected from an analysis of outliers (Box Plot). For comparative analysis between the predictor variables, we used general linear models (GLMs), which provide a comparison of categorical (predictors) and numerical (dependent) variables, and corrected for variables that may affect the outcome (covariates). The βS-haplotypes were selected as predictors; the hydroxycarbamide (HC) use, age, and genders as covariates; and the Hb F levels, hematological index, hemolytic index, and severity score as dependent variables. When appropriate, we applied the Fisher's post hoc test. Wilk’s λ test was performed as a multivariate analysis, which indicates the proportion of generalized variance in the dependent variables that is accounted for in the predictors. Data were expressed as mean ± 95% confidence interval and differences with a p value <0.05 were considered significant. Patients with the XmnI (rs7482144) polymorphism were not included in the statistical analysis due to the influence in the Hb F levels.
Results
Study group characterization
One thousand and two hundred chromosomes of SCA patients were classified as βS-haplotypes. Among the typical haplotypes, the most frequent was the Bantu haplotype (N = 879; 73.2%), followed by the Benin haplotype (N = 239; 19.9%). The Cameroon haplotype presented a frequency of 0.6% (N = 7) and the Saudi and Senegal presented the same frequency of 0.3% (N = 4). In the sample group, we found 63 (5.7%) patients with 11 haplotype patterns different from those previously described in the literature. They were named Atypical and classified according to observed frequency in Arabic numerals from 1 to 11, according to Fig. 1.
The most common atypical haplotype was Atypical 1 (N = 32; 47.8%), followed by Atypical 2 (N = 8; 12.0%) and 3 (N = 7; 10.4%). Atypical 4, 5, and 6 presented the same frequency (N = 4; 5.9%); Atypical 7 with 4.6% (N = 3); Atypical 8 with 3.0% (N = 2); and the Atypical 9, 10, and 11 were verified in just one chromosome each (1.5%). The criteria used to classify atypical haplotypes with the same frequency were based on the amount of polymorphic sites present (+) for the restriction enzymes. Genotype and haplotype profiles show that 55.2% SCA patients presented Bantu/Bantu, 24.5% Bantu/Benin, and 7.9% Bantu/Atypical haplotypes (Table 1).
We built a haplotype tree according to the presence (+) or absence (−) of restriction enzyme sites that characterized βS-haplotypes in order to understand the derivation of each atypical haplotype from the typical ones (Fig. 2). Bantu haplotypes might have originated four atypical haplotypes, the Benin three atypical haplotypes, the Cameroon and Senegal two atypical haplotypes, and the Saudi just one atypical haplotype. The majority of the 11 haplotype patterns described probably occurred by point mutation in typical and atypical haplotypes. A gene recombination event also may have resulted in atypical haplotypes. For example, Atypical 1 and 5 can be derived from a gene recombination between Bantu and Benin haplotypes, the Atypical 2 and 6 from Cameroon and Benin haplotypes, and the Atypical 3 and 9 from Bantu and Cameroon haplotypes. Senegal, Atypical 8 and 11 were not part of any branching haplotype tree.
Haplotype tree. HBG2: γ-glicin gene; HBG1: γ-alanin gene; HBBP: pseudo-β gene; HBB: β gene. (−) absence of enzyme restriction sites; (+) presence of enzyme restriction sites. Dark gray squares indicate the haplotypes that may have originated from gene recombination or point mutation. Light gray squares indicate the haplotypes that may have originated only by point mutation
Evaluation of study parameters
We compared the Hb F levels, hematological and hemolytic markers, and score of severity among haplotype groups to verify the haplotype influence in the studied parameters. The first group evaluated was composed of homozygous patients for the Bantu haplotype and Bantu/Atypical patients, including all the Bantu individuals with at least one atypical haplotype.
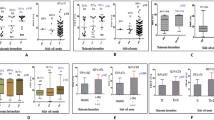
We verified that RBC levels in Bantu/Bantu patients were low compared to the same parameters in Bantu/Atypical patients (Fig. 3a, panels A1, A2 and A3). The variation found in hematological parameters was mostly (96%) explained by the haplotypes (Wilk’s λ: 0.96; F: 2.53; p value: 0.04). Thus, we showed a modulating effect of atypical haplotypes on hematological parameters.
Hematological index among sample groups: a Difference between Bantu/Bantu × Bantu/Atypical groups: (A1) red blood cells (RBC M/µL), (A2) hemoglobin (Hb, g/dL) and (A3) hematocrit (Ht, %). b Difference among Bantu/Bantu × Bantu/Atypical 1 × Bantu/Atypical* groups: (B1) red blood cells (RBC, M/μL), (B2) hemoglobin (Hb, g/dL) and (B3) hematocrit (Ht, %). c Difference among Bantu/Bantu × Bantu/Atypical 1 × Bantu/Atypical* × Benin/Benin × Benin/Atypical 1 groups: (C1) hemoglobin (Hb, g/dL) and (C2) hematocrit (Ht, %). Comparison made by general linear models. Data were expressed as mean ± 95% confidence intervals
A second analysis was performed among SCA patients with Bantu/Bantu, Bantu/Atypical 1, and Bantu/Atypical* haplotypes. We included in the Bantu/Atypical* group all the patients with atypical haplotypes associated with Bantu haplotypes, except the patients with Atypical 1 haplotype. Two criteria were essential for this second group formation: (1) Atypical 1 haplotype is the most frequent atypical in our study population providing a good sample size for statistical analysis; and (2) Atypical 1 does not present a restriction site for the enzymes used in the βS-haplotypes classification. After statistical analyses we verified that the patients with Bantu/Atypical 1 haplotypes showed the best values for RBC, Hb, and Ht levels compared to patients with Bantu haplotypes in homozygous form (Fig. 3b, panels B1, B2, and B3).
The comparison in the Benin group occurred between Benin/Benin and Benin/Atypical 1, because it was not possible to form the Benin/Atypical* group due to the small sample size. We did not find statistical differences in the hematological and hemolytic markers or clinical severity between the patients with Benin haplotypes in these two sample groups (data not shown).
We compared the five sample groups and verified that individuals with the Bantu haplotype in combination to Atypical 1 haplotype showed the best Hb and Ht values compared to Bantu homozygote patients (Hb p value: 0.002; Ht p value: 0.014; p value by Fisher's post hoc test) and Benin homozygote patients (Hb p value: 0.002; Ht p value: 0.016; p value by Fisher's post hoc test) (Fig. 3c, panels C1 and C2). Table 2 shows the mean and confidence interval of the parameters evaluated in the five groups.
Polymorphic sites that characterize βS globin haplotypes and TFs involved
We evaluated six restriction sites used to classify the βS-haplotypes in order to better understand the results obtained (Table 3). According to the β-globin cluster sequence available on the NCBI database, the combination formed (− + + − − + ) characterized the wild pattern haplotype. From the base substitutions, we know that in XmnI site occurs the exchange C → T in the promoter region in the HBG2 (−158pb position C → T; rs7482144); in the HincII site occurs the exchange G → A in 1140pb position and the exchange T → A in 2325pb position in the HBBP gene. Regarding the analysis of these three changes, it was possible to report four TF motifs that may influence the SCA phenotype: (1) TF CP2 (TFCP2) in mutated sequence in 5′-HBG2 region, (2) the MYB proto-oncogene (Myb) in mutated and wild sequences in HBBP region, (3) the GATA binding protein 1 and 2 factors (GATA-1 and GATA-2) to wild sequence in 3′HBBP region, and (4) the GATA-2 mutated sequence in 3′-HBBP region. In the other three restriction sites that compose the βS-haplotypes, it was not possible to identify the genetic changes in HindIII and HinfI enzyme sites.
Discussion
The point mutation responsible for SCA is not enough to explain the wide phenotypic variability found among individuals affected by the disease. Studies involving βS-haplotype, taking the approach of analyzing genetic modulators in SCA clinical severity, can be an useful tool to infer a severity profile, providing a more appropriate and effective treatment [12]. To our knowledge, this is the first research that suggests a classification for atypical haplotypes and checks possible modulation effects on Hb F levels, hematological and hemolytic parameters, and clinical severity in SCA patients.
Atypical haplotypes in our study population were the third most frequent after the Bantu and Benin haplotypes, with 5.7%. This frequency was expected because the atypical haplotypes are associated with 5 to 10% of the whole βS-chromosome [10]. We described 11 different patterns according to the presence or absence of six restriction sites. The description of several atypical haplotypes reflects the miscegenation of the study population as well as the high mutation rate in the cluster-β-globin. Studies showed that 3′ cluster-ββ regions, where HBBP, HBD, and HBB genes are located, is called a “hot spot” region because it is characterized by mutations occurring at a rate 100 times higher than normal [21, 22].
The atypical haplotypes origin occurs due to several genetic mechanisms involving the most common βS-haplotypes like Bantu and Benin. These mechanisms are: point mutations in typical haplotypes; simple and double recombination between a βS-chromosome either with a common βS-haplotype or with a less common haplotype associated with a βA-chromosome present in a given population; and, less frequently, gene conversion [6, 9, 10]. Out of a total of 63 chromosomes with atypical haplotypes, 47.8% were named Atypical 1 for being the most frequent. The Atypical 1 haplotype is characterized by the absence of restriction recognition sites, suggesting that Atypical 1 is a hybrid formed by gene recombination between 5ʹ Benin region and 3′ Bantu region. The other reason for the elevated frequency of Atypical 1 could be the loss of the restriction site for the enzyme HindIII in HBG2 gene in Bantu haplotype, or the loss of the restriction site for the enzyme HincII in 3′-HBBP gene in Benin haplotype by point mutations. The analysis of the haplotype tree drawn for the study population showed that approximately 91.0% of the atypical haplotypes are derived from point mutations in typical haplotypes.
The parameter analysis showed that SCA patients who were homozygous for Bantu haplotype presented worse rates of RBC, Hb, and Ht in relation to SCA patients with Bantu haplotype in interaction with any atypical haplotype. These results corroborate with the literature, which shows the relationship between the Bantu haplotype and the worst outcomes for inflammation, oxidative stress, and DNA damage [23,24,25,26].
In the comparison among Bantu/Bantu, Bantu/Atypical 1, and Bantu/Atypical*, we verified that Bantu/Atypical 1 patients showed better values of RBC, Hb, and Ht. With this result, we can suggest that the presence of Atypical 1 haplotype can contribute to a moderate clinical severity in SCA patients. This finding was corroborated when evaluating individuals with Benin haplotype. In the comparison made between Benin/Benin and Benin/Atypical 1 patients, we did not find a difference in any parameter. Benin haplotype causes in the patient a moderate pathophysiology compared to Bantu haplotype [11, 23, 24]. When comparing all sample groups, we found that the Atypical 1 haplotype in combination with Bantu contributes to better RBC, Hb, and Ht values in relation to individuals homozygous for Bantu and homozygous Benin. [27] We do not know what might have influenced the better values of these markers in patients with Atypical 1 haplotype, but we believe that the genotype that differs from Bantu presents a similar response to Benin haplotype.
Since the beginning of cluster β-globin studies, haplotypes are characterized by the presence or absence of restriction sites for specific enzymes [3, 5, 7, 8]. Considering that, mostly, the β-globin cluster is a “hot spot” region, any change in a nitrogenous base can contribute to the loss of restriction site for enzymes, as in the restriction site for HindI and HinfI. According to the β-globin cluster analysis in wild individuals, we did not find which nitrogenous base is changed in order to provide or not provide the recognition by the restriction enzyme in HindIII and HinfI sites. Thus, depending on the nitrogenous base changed, different TFs may act in the gene regulation, and probably in the phenotypic characteristics of the individual. In this way, we can explain why individuals with the same pattern of haplotypes presented differences between the same parameters or why individuals with different haplotypes do not present the expected phenotypic variation.
For XmnI and HincII restriction enzymes, four TFs that could act in the β-globin cluster gene regulation were listed. TF CP2 presented a relationship with cluster β-globin regulation in the presence of XmnI restriction sites (5′-HBG2-C → T, rs7482144). This factor activates the transcription of the α-globin gene (HBA) and indirectly regulates expression of HBG genes when it forms a complex with the specific factor of the erythrocyte (nuclear factor, erythrocyte 4) [28, 29]. The HincII enzyme recognizes a site in HBBP (G1140A) and another site in 3′-HBBP region (T2325A). In HBBP site (G1140A) the TF Myb presented a relationship with cluster β-globin regulation regardless of whether the sequence was wild or mutant. This factor regulates vertebrate hematopoiesis, but acts as a negative regulator of Hb F levels by inhibiting the expression of HBG [29,30,31]. In 3′-HBBP site (T2325A) of HincII, both GATA-1 and GATA-2 factors can interfere in the modulation of β-globin cluster of the wild sequence, and only the GATA-2 can interfere in the sequence mutated. GATA-1 factor is essential for the survival and final maturation of the erythrocytes precursors [32]. However, in the globin regulation it is involved in the silencing of HBG, because it forms Hb F protein repressor complexes [33]. GATA-2 factor forms complexes that activate the HBG genes transcription [34].
We believe that the βS-haplotypes modulate the SCA pathophysiology because different TFs can act according to the presence or absence of the restriction sites that characterize the βS-haplotypes. The four TFs reported above show direct or indirect relation with the expression regulation of the HBG genes. Myb, GATA-1, and GATA-2 factors, by being present in the HBBP gene region, which is a pseudo-gene, may not exhibit significant activity as genetic factors. Thus, to better understand the modulation of haplotypes in clinical severity of SCA patients, a more detailed study of the β-globin cluster is necessary to find and validate all possible TFs able to influence the phenotypic expression of SCA patients.
The present study suggested a classification for 11 atypical haplotypes patterns found in a population of unrelated SCA patients and most of the atypical haplotype seem to be derived from point mutations in typical haplotypes. The Atypical 1 haplotype is the most common and its presence was related to a mild phenotypic expression. A detailed study of restriction sites that characterize the βS-haplotypes and the identification of TFs that can influence the modulation of β-globin cluster may assist in understanding the pathophysiology found in SCA patients.
References
Rees DC, Williams TN, Gladwin MT. Sickle-cell disease. Lancet. 2010;376:2018–31.
Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–8.
Kan YW, Dozy M. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc Natl Acad Sci USA. 1978;75:5631–5.
Mears JG, et al. Its origin and diffusion from West Africa. J Clin Invest. 1981;68:606–10.
Pagnier J, et al. Evidence for the multicentric origin of the sickle cell hemoglobin gene in Africa. Proc Natl Acad Sci USA. 1984;81:1771–3.
Hockham C, Piel FB, Gupta S, Penman BS. Understanding the contrasting spatial haplotype patterns of malaria-protective beta-globin polymorphisms. Infect Genet Evol. 2015;36:174–83.
Kulozik aE, et al. Geographical survey of beta S-globin gene haplotypes: evidence for an independent Asian origin of the sickle-cell mutation. Am J Hum Genet. 1986;39:239–44.
Lapouméroulie C, et al. A novel sickle cell mutation of yet another origin in Africa: the Cameroon type. Hum Genet. 1992;89:333–7.
Srinivas R, et al. Atypical haplotypes linked to the beta-S gene in Africa are likely to be the product of recombination. Am J Hematol. 1988;29:60–62.
Zago Ma, et al. Atypical beta(s) haplotypes are generated by diverse genetic mechanisms. Am J Hematol. 2000;63:79–84.
Nagel RL. The origin of the hemoglobin S gene: clinical, genetic and anthropological consequences. Einst Q J Biol Med. 1984;2:53–62.
Gabriel A, Przybylski J. Sickle cell anaemia: a look at global haplotype distribution. Nat Educ. 2010;3:2.
Sambrook J, Fritsch EF, Maniatis, T MolecularCloning: a laboratory manual. Cold Spring Harb Lab Press 1989. http://www.molecularcloning.com/.
Saiki RK, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science (80-). 1985;230:1350–4.
Sutton M, Bouhassira EE, Nagel RL. Polymerase chain reaction amplification applied to the determination of beta-like globin gene cluster haplotypes. Am J Hematol. 1989;32:66–69.
Nouraie M, et al. The relationship between the severity of hemolysis, clinical manifestations and risk of death in 415 patients with sickle cell anemia in the US and Europe. Haematologica. 2013;98:464–72.
Sebastiani P, et al. A network model to predict the risk of death in sickle cell disease. Blood. 2007;110:2727–35.
Belini Junior E, et al. Severity of Brazilian sickle cell disease patients: severity scores and feasibility of the Bayesian network model use. Blood Cells Mol Dis. 2015;54:321–7.
Mathelier A, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:142–7.
Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4.
Hanchard N, et al. Classical sickle beta-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genet. 2007;8:52.
Chakravarti a, et al. Nonuniform recombination within the human beta-globin gene cluster. Am J Hum Genet. 1984;36:1239–58.
Powars DR. Beta s-gene-cluster haplotypes in sickle cell anemia. Clinical and hematologic features. Hematol Oncol Clin N Am. 1991;5:475–93.
Powars D, Hiti A. Sickle cell anemia. Beta s gene cluster haplotypes as genetic markers for severe disease expression. Am J Dis Child. 1993;147:1197–202.
Silva-Rocha LB, Dias-Elias DB, Barbosa MC, Bandeira ICJ, Gonçalves RP. DNA damage in leukocytes of sickle cell anemia patients is associated with hydroxyurea therapy and with HBB*S haplotype. Mutat Res Toxicol Environ Mutagen. 2012;749:48–52.
Steinberg MH, et al. Fetal hemoglobin in sickle cell anemia: determinants of response to hydroxyurea. Multicenter Study of Hydroxyurea. Blood. 1997;89:1078–88.
Nagel RL, et al. The hematologic characteristics of sickle cell anemia bearing the Bantu haplotype: the relationship between G-y and Hb F levels. Blood. 1987;69:1026–30.
Zhou W, et al. The role of p22 NF-E4 in human globin gene switching. J Biol Chem. 2004;279:26227–32.
Carrocini GC, de S, Venancio LPR, Bonini-Domingos CR. Screening of transcription factors involved in fetal hemoglobin regulation using phylogenetic footprinting. Evol Bioinforma. 2015;11:239–44.
Soza-ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci USA. 2010;107:17340–08.
Bauer D, Kamram S, Orkim S. Reawakening fetal hemoglobin: prospects for new therapies for the β-globin disorders. Blood. 2012;120:2945–53.
Ikonomi P, et al. Levels of GATA-1/GATA-2 transcription factors modulate expression of embryonic and fetal hemoglobins. Gene. 2000;261:277–87.
Xu J, et al. Transcriptional silencing of beta-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–9.
Zhu X et al. NF-Y recruits both transcription activator and repressor to modulate tissue- and developmental stage-specific expression of human y-globin gene. PLoS ONE 2012;7: 1–15.
Acknowledgements
We would like to thank Izabella Luisa Tambones (Department of Biology, São Paulo State University – UNESP) for her technical assistance in transcription factors analyses and Joseph People (Department of Physiology, Development and Neuroscience, University of Cambridge) for English proofreading. The Pro-Institute of Hematology Foundation (Fundarj) and the State of Sao Paulo Research Foundation (FAPESP process number 2011/14168-5) for the financial support. Silva is a partnership member of Redoxoma – CEPID/FAPESP (2013/07937-8).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Okumura, J.V., Silva, D.G.H., Torres, L.S. et al. Atypical β-S haplotypes: classification and genetic modulation in patients with sickle cell anemia. J Hum Genet 64, 239–248 (2019). https://doi.org/10.1038/s10038-018-0554-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0554-4