Abstract
Nail-patella syndrome (NPS) is an autosomal dominant disease characterized by dysplastic nails, absent or hypoplastic patellae, elbow dysplasia, and nephropathy. Recently, it was shown that NPS is the result of heterozygous mutations in the LIM-homeodomain gene, LMX1B. Subsequently, many mutations of the LMX1B gene have been reported in NPS patients. However, functional analyses of the mutant proteins have been performed in only a few mutations. Furthermore, the mechanisms of dominant inheritance in humans have not been established. In the present study, we analyzed the LMX1B gene in three Japanese patients with NPS and identified two novel mutations, 6 nucleotide deletion (Δ246Ν 247Q) and V242L. These two mutations are located in the homeodomain of LMX1B. Functional analyses of the LMX1B mutants revealed that these mutants had diminished transcriptional activity and had lost DNA binding ability. Furthermore, we demonstrated that each mutant did not manifest a dominant-negative effect on the transcriptional activity of wild-type LMX1B. These results suggested that NPS is caused by loss-of-function mutations of LMX1B, and haploinsufficiency of LMX1B should be the predominant pathogenesis of NPS in humans.
Similar content being viewed by others
Main
Nail-patella syndrome (NPS; MIM#161200) is an autosomal dominant disease characterized by dysplastic nails, absent or hypoplastic patellae, elbow dysplasia, iliac horns, and, in some cases, open-angle glaucoma and nephropathy (1–3). Nail dysplasia is the most constant feature of NPS observed at birth. The most serious aspect of NPS is nephropathy, which might develop end-stage renal failure. Although development to end-stage renal failure is usually slow, a minority of cases have been reported to show rapid progression during early childhood (3,4).
It has been demonstrated that mutations of the LMX1B gene, which is located on chromosome 9q34, result in NPS (5–8). LMX1B is a member of the LIM-homeodomain family of transcription factors that are involved in body-pattern formation during development (9,10). These proteins contain two cysteine-rich zinc-binding motifs (LIM-A and LIM-B domain) at their amino termini that are important in mediating protein–protein interactions and a homeodomain involved in DNA binding (9,10). The LMX1B-mediated transactivation presumably requires interaction with a transcriptional complex including a helix-loop-helix protein, E47/shPan1 (11,12). During embryogenesis, Lmx1b is strongly expressed in dorsal mesenchymal tissues in mice (12). Lmx1b−/− mice showed the absence of dorsal limb structures and duplication of ventral structures (7). These findings suggest that Lmx1b is essential for dorsoventral patterning during development and that the skeletal phenotype of NPS is the result of a deficiency in dorsoventral patterning. LMX1B is also expressed in human fetal and adult kidneys (5).
From the analysis of Lmx1b−/− mice, it was suggested that LMX1B regulates the expression of type IV collagen α3 and α4 and podocin and that Lmx1b mutations cause disruption of these proteins, resulting in abnormal glomerular basement membrane morphogenesis and podocyte differentiation, leading to nephropathy (13,14). However, in contrast to the mouse model for NPS, the expression of type IV collagen α3 and α4 and podocin was normal in patients with NPS (15). LMX1B has also been shown to be essential for the development of serotonergic neurons in the CNS (16–18).
The human LMX1B gene consists of eight exons spanning a region of ∼82 kb (8). The transcript is ∼7 kb in length and is predicted to encode a protein of 372 amino acid residues (5). To date, >80 different heterozygous mutations of the LMX1B gene have been reported in patients with NPS (4). The mutations are concentrated within the LIM domains (LIM-A 44%, LIM-B 38%) and the homeodomain (18%) and exist exclusively in exons 2–5 (4). The most frequently recurring mutations have been found in the homeodomain (R198X, R200Q, R208X, A213P, and R223X), which comprises nearly 30% of all known LMX1B mutations. However, no mutations in the LMX1B gene were detected in 19% of patients with NPS (4). In most cases reported previously, Southern blotting analysis was not performed and the possibility of large genomic rearrangements in NPS gene could not be excluded (19).
Although a large number of LMX1B mutations had been reported, functional analyses of mutant proteins had been performed in only five mutations (5,12). Dreyer et al. (12) showed that transcriptional activities of the four missense mutants in the homeodomain were eliminated, whereas one in the LIM-B domain had residual activity. Because of limited information on functional characterization of mutant LMX1B proteins, the mechanism of dominant inheritance in humans has not been established. Although haploinsufficiency mechanism of LMX1B in heterozygous LMX1B mutations had been suggested, the possibility of dominant-negative effect has not been ruled out, especially because deletions of the entire gene have not been reported.
In this report, we performed genetic analysis in LMX1B gene in patients with NPS and identified two novel mutants. We conducted functional analyses of the mutant proteins detected in the patients to elucidate the transcriptional activity and DNA binding of these mutant proteins. Furthermore, we investigated the dominant-negative effect of LMX1B mutants.
METHODS
Patients.
Patient 1 was a 3-y-old Japanese girl with proteinuria and hematuria. Urine abnormalities were pointed out in the urinary examination program as a 3-y-old. Her physical examination revealed absent nails in thumb and index fingers and hypoplastic nails in others. She manifested limited extension and supination of the elbow joints. Her laboratory data were as follows. Urine analysis revealed mild hematuria, mild proteinuria, and glucosuria. Serum chemistry showed total protein of 6.7 g/dL, albumin of 3.5 g/dL, blood urea nitrogen of 7 mg/dL, creatinine of 0.2 mg/dL, uric acid of 3.2 mg/dL, sodium of 141 mEq/L, potassium of 4.4 mEq/L, chloride of 105 mEq/L, calcium of 9.8 mg/dL, phosphate of 4.8 mg/dL, and glucose of 108 mg/dL. Her radiologic examinations revealed iliac horns and absent patellae. Glaucoma was not detected. Her paternal grandfather died from renal insufficiency at 35 y of age, and her father and a paternal uncle had proteinuria and hypoplastic nails.
Patient 2 was a 16-y-old Japanese boy with hypoplastic thumbnails. He was referred to our hospital because of anorexia nervosa. Radiologic examinations revealed absent patellae and presence of iliac horns. His elbow joint was normal, and glaucoma was not detected. His laboratory data were as follows. Serum chemistry showed total protein of 7.2 g/dL, blood urea nitrogen of 10 mg/dL, and creatinine of 0.6 mg/dL. His urinalysis was normal, and he had no signs of nephropathy. His father showed similar clinical features and had received a diagnosis of NPS. His father also did not develop nephropathy.
Patient 3 was a 14-y-old Japanese boy with proteinuria detected by a urinary screening program in school children. He had hypoplastic nails and limited extension of elbows. Radiologic examinations revealed absent patella and iliac horns. The iris nodules were indicated in his angulus iridocornealis by an ophthalmologist. His laboratory data were as follows. Urine analysis revealed severe proteinuria (urine protein 1.5–2.5 g/d) and no hematuria. Serum chemistry showed total protein of 5.6 g/dL, albumin of 3.3 g/dL, blood urea nitrogen of 14 mg/dL, and creatinine of 0.53 mg/dL. His creatinine clearance was 129 mL/min. Renal biopsy was performed. Minimal glomerular change was detected with light microscopy, and electron microscopy examination revealed specific histologic findings of NPS, including irregular thickening of the glomerular basement membrane that contained patchy electron-lucent areas (“moth-eaten appearance”) and fibers with striated collagen periodicity. Neither of his parents had symptoms of NPS.
LMX1B gene analysis.
Informed consent for DNA analysis was obtained from the patients and parents. The study was performed with the consensus of the ethical committee of the Tokyo University. Genomic DNA was extracted from peripheral white blood cells of all of the patients and the parents of patient 3 by proteinase K digestion and phenol/chloroform extraction. PCR was performed to amplify the entire coding region and exon-intron boundaries of the LMX1B gene. The primers used for PCR amplification were as follows: exon 1 sense 5′-GGCAGACGGACTGCGCC-3′, exon 1 antisense 5′-GTGTCCACAGCCGGACGAC-3′, exon 2 sense 5′-GGCCCGGTGCGACCGGGAC-3′, exon 2 antisense 5′-GCTGACCGGGCTCGAGTGC-3′, exon 5–6 sense 5′-GGTAGGGACATCCCTCCA-3′, exon 5–6 antisense 5′-CCAGCTCACCCTGGCCTAGG-3′, and as described for exons 3, 4, 7, and 8 (6). Exons 1, 3, 4, 5–6, and 8 were amplified in a final volume of 50 μL, using 0.4 μM of each primer, 100 ng of genomic DNA, 5 μL of GeneAmp 10× DNA PCR Buffer, 0.08 mM dNTP, and 2.5 units of AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA), with a PCR condition of 94°C for 9 min, 35 cycles of 94°C for 30 s, 62°C for 30 s, 72°C for 1 min, followed by 7 min at 72°C (20). Exons 2 and 7 were amplified in a final volume of 50 μL, using 0.8 μM of each primer, 100 ng of genomic DNA, 5 μL of 10× Cloned Pfu DNA polymerase reaction buffer, 0.08 mM dNTP, and 3.75 units of Pfu Turbo DNA Polymerase (Stratagene, La Jolla, CA), with a PCR condition of 98°C for 3 min, 35 cycles of 98°C for 30 s, 60°C for 30 s, 72°C for 1 min, followed by 7 min at 72°C. The PCR products of exons 1, 3, and 8 were purified using a PCR purification kit (Qiagen, Hilden, Germany), and the PCR products of exons 2, 4, 5–6, and 7 were electrophoresed on agarose gel and purified using a gel purification kit (Qiagen). Direct sequencing in both directions was performed on an autosequencer (ABI PRISM 310, Genetic Analyzer; Applied Biosystems). Detected mutations were confirmed by cloning the PCR products into the pCR2.1 vector using a TOPO TA Cloning kit (Invitrogen, Carlsbad, CA), and the clones derived from both alleles were sequenced.
For Southern blot analysis, 10 μg of genomic DNA was digested with restriction enzyme PstI and was subjected to electrophoresis on 0.8% agarose gels, transferred to charged nylon filters (Amersham Pharmacia Biotech, Piscataway, NJ), and hybridized with DNA probes. Probes were generated by a PCR DIG Probe Synthesis Kit (Roche Applied Science, Indianapolis, IN) using wild-type LMX1B cDNA as a template and primers for amplifying probe A (1–489 bp, involving exons 1–3) or probe B (508–750 bp involving exons 4–5) or probe C (712–1125 bp involving exons 6–8). Hybridization, washing, blocking, and detection of the bands were performed using DIG Easy Hyb, DIG Wash and Block Set, DIG Luminescent Detection Kit (Roche Applied Science) according to the manufacturer's protocol. We also performed a similar experiment using probe A and genomic DNA digested with XbaI.
Construction of plasmids.
The constructs of wild-type LMX1B expression plasmid LMX1B-pcDNA3.1, shPan1 expression plasmid shPan1-pBAT14, and luciferase reporter plasmid 5xFar/FLAT-pFOXluc1.prl that contained five copies of the rat insulin mini-enhancer element E2A3/4(Far1/FLAT) were described previously (11,12,21). Mutations Δ246Ν247Q and V247L in the LMX1B expression plasmid were introduced with a Quick Change Site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Each LMX1B mutant construct was sequence-verified to have no extra mutations.
Cell culture and transfection.
COS-7 and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in humidified 95% air and 5% CO2 at 37°C. Cells that were cultured in 24-well plates were transfected with 250 ng of DNA, including 100 ng of the reporter gene, 10 ng of shPAN1-pBAT14, the indicated amount of each LMX1B expression plasmids, and 0.1 ng of pRL-CMV (Promega, Madison, WI) using LipofectAMINE Plus (Invitrogen) (22). After 24 h, the transcriptional activity was assayed using a Dual-Luciferase Reporter Assay System (Promega). The luciferase activities of 5xFar/FLAT-pFOXluc1.prl were normalized to the luciferase activities of pRL-CMV.
In vitro translation and electrophoretic mobility shift analysis.
LMX1B-wild type, LMX1B-Δ246N,247Q, LMX1B-V247L protein were synthesized with wild-type LMX1B-pcDNA3.1, LMX1BΔ246N,247Q-pcDNA3.1, LMX1BV247L-pcDNA3.1 using the TNT T7 Coupled Reticulocyte Lysate System (Promega). Equivalent amounts of proteins, as determined by SDS-PAGE and autoradiography of [35S]methionine-labeled proteins, were incubated with a 32P-labeled double-stranded oligonucleotide that contained the insulin mini-enhancer sequence as described (22). The DNA-protein complexes were resolved on a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate, EDTA buffer. The gel then was dried and exposed to x-ray film for autoradiography.
RESULTS
Mutation analysis of the LMX1B gene.
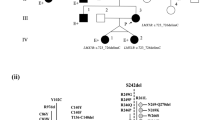
For investigating mutations in the LMX1B gene, all exons of LMX1B gene in the three patients were PCR-amplified and sequenced directly. Detected mutations were confirmed by repeated PCR and sequencing in both directions, as well as by sequencing the subcloned PCR products. Patient 1 had a heterozygous 6-bp deletion in exon 5 (Fig. 1A). This 6-bp deletion is predicted to result in the deletion of asparagine and glutamine in codon 246–247 (Δ246Ν247Q). Patient 3 had heterozygous G to C transition in exon 5 (Fig. 1B). This transition is predicted to result in substitution of valine to leucine in codon 242 (V242L). This V242L mutation was not found in the father or the mother of patient 3, suggesting a de novo mutation. The absence of each DNA sequence abnormality in 100 alleles from 50 unrelated normal Japanese individuals established that these abnormalities were mutations and not a polymorphism that would be expected to occur in >1% of the population. In patient 2, no mutation was found in any exons or exon-intron boundaries in the LMX1B gene. To investigate larger rearrangements in the LMX1B gene, we performed Southern blot analysis of the LMX1B gene in patient 2. PstI digestion of genomic DNA of patient 2 and hybridization with the probes identified bands of 2.3- and 1.5-kb fragment for probe A, 1.5-kb fragment for probe B, and 2.5- and 1.6-kb fragment for probe C, respectively (data not shown). The fragment sizes were similar to those detected in normal control subjects. To confirm the result, we performed additional Southern blot analysis using probe A with XbaI digestion of genomic DNA of patient 2. Hybridization with probe A identified 14.6-, 9.2-, 8.6-, and 6.2-kb fragment. The fragment sizes were also similar to those detected in normal control subjects. These data suggested that this patient did not have large rearrangements in the LMX1B gene.
Δ246Ν247Q and V242L mutations in the LMX1B gene identified in patient 1 and patient 3. (A) Patient 1 had a heterozygous 6-bp deletion in exon 5 predicted to result in the deletion of 246Asn and 247Gln (Δ246Ν247Q). (B) Patient 3 had a heterozygous G to C transition in exon 5 predicted to result in the substitution of Val to Leu in codon 242 (V242L). The top panel shows the sequencing chromatograms obtained from a normal control, the middle panel shows the direct sequencing of the patient, and the bottom panel shows the subcloned mutant allele.
Transcriptional activity of LMX1B mutants.
For examining whether both mutations may affect LMX1B transactivation, wild-type LMX1B and mutant LMX1B were transiently overexpressed in mammalian cells, and the transcriptional activity was analyzed by luciferase reporter assay. We used a mini-enhancer of the rat insulin I gene, E2A3/4 (Far/FLAT) sequence, which has been known to be bound and transactivated by LMX1B (12). Transfection of the wild-type LMX1B efficiently increased the reporter gene activity in a dose-dependent manner (Fig. 2). However, both mutants had negligible transcriptional activity. Similar results were also observed in HeLa cells (data not shown). Therefore, LMX1B-Δ246N,247Q and LMX1B-V242L mutants had diminished transcriptional activity.
The transcriptional activity of LMX1B-Δ246N,247Q and LMX1B-V242L. The indicated amounts of each LMX1B expression plasmid were transfected with 100 ng of the insulin mini-enhancer reporter into COS-7 cells. The relative luciferase activity (Firefly/Runilla) was measured by three independent experiments. Fold induction refers to the activity without any LMX1B. The mean ± SD is shown. All differences between the corresponding dose of the mutants and wild-type were significant (p < 0.05).
DNA binding ability of the LMX1B mutants.
We further examined whether the diminished transcriptional activity of LMX1B-Δ246N,247Q and LMX1B-V242L was due to the loss of DNA binding. The transcriptional efficiency was checked by SDS-PAGE analysis, which indicated that proteins of the expected sizes (42 kD) were synthesized (Fig. 3A). Electrophoretic mobility shift analysis (EMSA) showed that wild-type LMX1B bound to the insulin mini-enhancer element, and this binding could be blocked by the addition of excess unlabeled oligonucleotide (Fig. 3B, lanes 2 and 3). However, LMX1B-Δ246N,247Q and LMX1B-V242L did not bind to the insulin mini-enhancer element (Fig. 3B, lanes 4 and 5). These data suggested that diminished transcriptional activity of these mutants was due to their inability to bind DNA.
DNA binding activity of the LMX1B-Δ246N,247Q and LMX1B-V242L. (A) In vitro translated products of pcDNA3 vector, pcDNA3-LMX1B-wild type, pcDNA-LMX1B-Δ246N,247Q, and pcDNA-LMX1B-V242L in the presence of [35S]methionine. (B) EMSA with the oligonucleotide probes corresponding to the rat insulin I mini-enhancer element. Unlabeled in vitro translated products were incubated with 32P-labeled DNA fragments and separated on a 5% nondenaturing polyacrylamide gel. The formation of the wild-type LMX1B binding complex was inhibited by adding excess cold probes (lane 3).
Dominant-negative effect of the mutants on wild-type LMX1B.
Because these mutations were detected heterozygously in both patients, we next examined whether these mutant LMX1B have a dominant-negative effect on wild-type LMX1B. Wild-type LMX1B was transfected with increasing amounts of LMX1B-Δ246N,247Q or LMX1B-V242L in COS-7 cells. The transcriptional activity of wild-type LMX1B was not repressed by co-transfection of LMX1B-Δ246N,247Q or LMX1B-V242L (Fig. 4A). Similar results were also observed in HeLa cells (data not shown). The effect of the mutants on wild-type LMX1B DNA binding was also analyzed by EMSA. Neither of the mutants inhibited wild-type DNA binding in the insulin mini-enhancer element (Fig. 4B). These results suggested that neither LMX1B-Δ246N,247Q nor LMX1B-V242L has an inhibitory effect on wild-type LMX1B transactivation and DNA binding.
Analysis of the dominant-negative effect of Δ246N,247Q and V242L on wild-type LMX1B transcriptional activity and DNA binding activity. (A) The indicated amounts of pcDNA-LMX1B-Δ246N,247Q or pcDNA-LMX1B-V242L were co-transfected with 20 ng of pcDNA-LMX1B-wild type and 100 ng of the insulin mini-enhancer reporter into COS-7 cells. The relative luciferase activity (Firefly/Runilla) was measured by three independent experiments. Fold induction refers to the activity with 20 ng of pcDNA-LMX1B-wild type. The mean ± SD is shown. All differences between the mutants and the control were not significant. (B) EMSA with oligonucleotide probes corresponding to the rat insulin I mini-enhancer element. Unlabeled in vitro translated products were incubated with 32P-labeled DNA fragments and separated on a 5% nondenaturing polyacrylamide gel. Equal amounts of LMX1B-Δ246N,247Q or LMX1B-V242L mutant did not inhibit wild-type LMX1B DNA binding.
DISCUSSION
We found two novel mutations, Δ246N,247Q and V242L, in the LMX1B gene in two patients with NPS. 242V, 246N, and 247Q are located in the third helix of the homeodomain (9,12), which is involved in DNA binding (9). Moreover, 242V, 246N, and 247Q are well conserved among human LMX1B, mouse Lmx1b, Xenopus Lmx1b, and human LMX1A. Thus, it is suggested that these amino acids are critical for LMX1B function and that both the Δ246N,247Q mutation and the V242L mutation are predicted to decrease the transactivity of LMX1B by disturbed DNA binding ability.
To confirm the function of the mutant LMX1B proteins, we analyzed the transcriptional activity and DNA binding ability of the two newly identified LMX1B mutants. Functional analysis revealed that both Δ246N,247Q and V242L mutations abolished the transcriptional activity as well as the DNA binding ability. Therefore, both LMX1B-Δ246N,247Q and LMX1B-V242L were loss-of-function mutations and these mutations are likely to cause NPS. Only one study has reported that four LMX1B mutations in the homeodomain abolish transcriptional activity, whereas one in the LIM-B domain had residual transcriptional activity (12). Here we identified that Δ246N,247Q and V242L in the homeodomain had no transcriptional activity and no DNA binding ability. These data suggest that the homeodomain is indispensable for full transcriptional activity and DNA binding of LMX1B, although some decrement of transcriptional activity may result in NPS.
Although it is clear that heterozygous LMX1B mutations cause NPS in humans and Lmx1b heterozygous null mice have no abnormal phenotype, the mechanism of the difference is unknown. This might be due to the difference in the species, and NPS might be caused by haploinsufficiency of LMX1B in humans. However, because deletions of the entire LMX1B gene have not been reported in NPS patients, the possibility of a dominant-negative effect of the mutant LMX1B proteins cannot be ruled out. Dreyer et al. (12) found some negative effects on wild-type LMX1B transactivation in some mutants. However, the transfected dose of wild-type DNA was not the same in their mixed study. In this study, we investigated the dominant-negative effect of LMX1B mutants by transfecting increasing amounts of the mutants with a fixed dose of wild-type LMX1B. We chose the dose of the wild-type that had intermediate transcriptional activity (Fig. 2) to detect both activation and repression effects. We found that neither LMX1B-Δ246N,247Q nor LMX1B-V242L affects the transcriptional activity of wild-type LMX1B using an insulin mini-enhancer element. We further found that these mutants have no effect on DNA binding of the wild-type LMX1B. Although there still is the possibility of a dominant-negative effect on other target gene promoters, these results suggested that NPS is caused by haploinsufficiency of LMX1B.
Genotype-phenotype correlation is not shown in patients with NPS (4). Moreover, there is marked intrafamilial variability of renal disease among members with the same LMX1B mutation (4,19,23,24). However, for assessing the correlation between the phenotype and functional abnormality, the number of mutations that have been analyzed is too small. Our functional analysis revealed that two novel mutant LMX1B proteins totally abolished transcriptional activity in families with severe renal disease. For clarifying whether the complete loss-of-function mutations in LMX1B are a risk factor to induce nephropathy, further functional analysis of the other LMX1B mutants will be necessary.
In patient 2, we found no mutation in the LMX1B gene by PCR direct sequencing and Southern blot analysis. Abnormality in the introns, the promoter region, or the 3′ untranslated region of LMX1B is also possible. Otherwise, mutations may be in other genes that are closely related to LMX1B, including the transcriptional co-factors.
Although we and others have clarified that loss-of-function heterozygous mutation is the cause of NPS, little is known about the molecular mechanism underlying the phenotype of NPS. Some candidate target genes of transcriptional factor LMX1B have been reported, such as type IV collagen α3 and α4 and podocin in the kidney (13,14). However, it is reported that expression of these proteins in NPS patients was normal, and it is not known whether abnormalities in these genes actually contribute to nephropathy in NPS (15). Moreover, little is known about genes upstream and downstream of LMX1B in other LMX1B-expressing organs. The mechanism underlying the intrafamilial phenotypic variations also remains to be clarified. For understanding these points and to identify factors that lead to nephropathy and glaucoma, further molecular studies may be necessary.
CONCLUSION
In summary, we identified two novel mutations of the LMX1B gene in Japanese patients with NPS. These LMX1B mutants had diminished transcriptional activity and DNA binding ability. Furthermore, these mutants did not have a dominant-negative effect on wild-type transcriptional activity. It is suggested that NPS is caused by haploinsufficiency of LMX1B.
Abbreviations
- EMSA:
-
electrophoretic mobility shift analysis
- NPS:
-
nail-patella syndrome
References
Beals RK, Eckhardt AL 1969 Hereditary onycho-osteodysplasia (Nail-Patella syndrome). A report of nine kindreds. J Bone Joint Surg Am 51: 505–516
Lichter PR, Richards JE, Downs CA, Stringham HM, Boehnke M, Farley FA 1997 Cosegregation of open-angle glaucoma and the nail-patella syndrome. Am J Ophthalmol 124: 506–515
Sweeney E, Fryer A, Mountford R, Green A, McIntosh I 2003 Nail patella syndrome: a review of the phenotype aided by developmental biology. J Med Genet 40: 153–162
Bongers EM, Gubler MC, Knoers NV 2002 Nail-patella syndrome. Overview on clinical and molecular findings. Pediatr Nephrol 17: 703–712
Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, Lee B 1998 Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet 19: 47–50
Vollrath D, Jaramillo-Babb VL, Clough MV, McIntosh I, Scott KM, Lichter PR, Richards JE 1998 Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail-patella syndrome. Hum Mol Genet 7: 1091–1098
Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg KC, Pepicelli CV, Gan L, Lee B, Johnson RL 1998 Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat Genet 19: 51–55
Iannotti CA, Inoue H, Bernal E, Aoki M, Liu L, Donis-Keller H, German MS, Permutt MA 1997 Identification of a human LMX1 (LMX1.1)-related gene, LMX1.2: tissue-specific expression and linkage mapping on chromosome 9. Genomics 46: 520–524
Curtiss J, Heilig JS 1998 DeLIMiting development. Bioessays 20: 58–69
Hobert O, Westphal H 2000 Functions of LIM-homeobox genes. Trends Genet 16: 75–83
German MS, Wang J, Chadwick RB, Rutter WJ 1992 Synergistic activation of the insulin gene by a LIM-homeo domain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev 6: 2165–2176
Dreyer SD, Morello R, German MS, Zabel B, Winterpacht A, Lunstrum GP, Horton WA, Oberg KC, Lee B 2000 LMX1B transactivation and expression in nail-patella syndrome. Hum Mol Genet 9: 1067–1074
Morello R, Zhou G, Dreyer SD, Harvey SJ, Ninomiya Y, Thorner PS, Miner JH, Cole W, Winterpacht A, Zabel B, Oberg KC, Lee B 2001 Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet 27: 205–208
Miner JH, Morello R, Andrews KL, Li C, Antignac C, Shaw AS, Lee B 2002 Transcriptional induction of slit diaphragm genes by Lmx1b is required in podocyte differentiation. J Clin Invest 109: 1065–1072
Heidet L, Bongers EM, Sich M, Zhang SY, Loirat C, Meyrier A, Broyer M, Landthaler G, Faller B, Sado Y, Knoers NV, Gubler MC 2003 In vivo expression of putative LMX1B targets in nail-patella syndrome kidneys. Am J Pathol 163: 145–155
Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP 2000 A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci 3: 337–341
Kania A, Johnson RL, Jessell TM 2000 Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell 102: 161–173
Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF 2003 Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci 6: 933–938
Clough MV, Hamlington JD, McIntosh I 1999 Restricted distribution of loss-of-function mutations within the LMX1B genes of nail-patella syndrome patients. Hum Mutat 14: 459–465
Kitanaka S, Takeyama K, Murayama A, Sato T, Okumura K, Nogami M, Hasegawa Y, Niimi H, Yanagisawa J, Tanaka T, Kato S 1998 Inactivating mutations in the 25-hydroxyvitamin D3 1α-hydroxylase gene in patients with pseudovitamin D-deficiency rickets. N Engl J Med 338: 653–661
German M, Ashcroft S, Docherty K, Edlund H, Edlund T, Goodison S, Imura H, Kennedy G, Madsen O, Melloul D, Serup P, Moss L, Olson K, Robertson P, Permutt A, Philippe J, Stein R, Steiner D, Tsai J, Walker D, Rutter J 1995 The insulin gene promoter. A simplified nomenclature. Diabetes 44: 1002–1004
Kitanaka S, Miki Y, Hayashi Y, Igarashi T 2004 Promoter-specific repression of hepatocyte nuclear factor (HNF)-1β and HNF-1α transcriptional activity by an HNF-1β missense mutant associated with type 5 maturity-onset diabetes of the young with hepatic and biliary manifestations. J Clin Endocrinol Metab 89: 1369–1378
McIntosh I, Dreyer SD, Clough MV, Dunston JA, Eyaid W, Roig CM, Montgomery T, Ala-Mello S, Kaitila I, Winterpacht A, Zabel B, Frydman M, Cole WG, Francomano CA, Lee B 1998 Mutation analysis of LMX1B gene in nail-patella syndrome patients. Am J Hum Genet 63: 1651–1658
Knoers NV, Bongers EM, van Beersum SE, Lommen EJ, van Bokhoven H, Hol FA 2000 Nail-patella syndrome: identification of mutations in the LMX1B gene in Dutch families. J Am Soc Nephrol 11: 1762–1766
Acknowledgements
We thank Dr. Brendan Lee and Dr. Michael S. German for providing the plasmids and Dr. Yu Yan Chen and Dr. Kojiro Sato for helpful advice and assistance in the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by Grant-in-Aid from the Ministry of Health and Welfare of Japan and from the Ministry of Education, Science, Sports, and Culture of Japan.
Rights and permissions
About this article
Cite this article
Sato, U., Kitanaka, S., Sekine, T. et al. Functional Characterization of LMX1B Mutations Associated with Nail-Patella Syndrome. Pediatr Res 57, 783–788 (2005). https://doi.org/10.1203/01.PDR.0000157674.63621.2C
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/01.PDR.0000157674.63621.2C
This article is cited by
-
Simultaneous kidney and pancreas transplantation in a patient with nail-patella syndrome and insulin-dependent diabetes
Pediatric Nephrology (2023)
-
Clinical and genetic characterization of nephropathy in patients with nail-patella syndrome
European Journal of Human Genetics (2020)
-
A novel small deletion of LMX1B in a large Chinese family with nail-patella syndrome
BMC Medical Genetics (2019)
-
Heterozygous missense variants of LMX1A lead to nonsyndromic hearing impairment and vestibular dysfunction
Human Genetics (2018)
-
Spectrum of LMX1B mutations: from nail–patella syndrome to isolated nephropathy
Pediatric Nephrology (2017)