Abstract
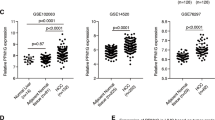
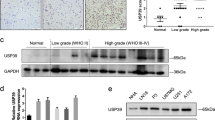
CD44E is a frequently overexpressed variant of CD44 in gastric cancer. Mechanisms that regulate CD44 splicing and expression in gastric cancer remain unknown. Herein, we investigated the role of DARPP-32 (dopamine and cyclic adenosine monophosphate-regulated phosphoprotein, Mr 32000) in promoting tumor growth through regulation of CD44 splicing. Using western blot and quantitative real-time PCR analysis, our results indicated that knockdown of endogenous DARPP-32 markedly reduces the expression of CD44 V8-V10 (CD44E). Using a quantitative splicing luciferase reporter system, we detected a significant increase in the reporter activity following DARPP-32 overexpression (P<0.001). Conversely, knocking down endogenous DARPP-32 significantly attenuated the splicing activity (P<0.001). Further experiments showed that DARPP-32 regulates the expression of SRp20 splicing factor and co-exists with it in the same protein complex. Inhibition of alternative splicing with digitoxin followed by immunoprecipitation and immunoblotting indicated that DARPP-32 has an important role in regulating SRp20 protein stability. The knockdown of endogenous DARPP-32 confirmed that DARPP-32 regulates the SRp20-dependent CD44E splicing. Using tumor xenograft mouse model, knocking down endogenous DARPP-32 markedly reduced SRp20 and CD44E protein levels with a decreased tumor growth. The reconstitution of SRp20 expression in these cells rescued tumor growth. In addition, we also demonstrated frequent co-overexpression and positive correlation of DARPP-32, SRp20 and CD44E expression levels in human gastric primary tumors. Our novel findings establish for the first time the role of DARPP-32 in regulating splicing factors in gastric cancer cells. The DARPP-32–SRp20 axis has a key role in regulating the CD44E splice variant that promotes gastric tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Mukherjee K, Peng D, Brifkani Z, Belkhiri A, Pera M, Koyama T et al. Dopamine and cAMP regulated phosphoprotein MW 32 kDa is overexpressed in early stages of gastric tumorigenesis. Surgery 2010; 148: 354–363.
El-Rifai W, Smith MF Jr, Li G, Beckler A, Carl VS, Montgomery E et al. Gastric cancers overexpress DARPP-32 and a Novel Isoform, t-DARPP. Cancer Res 2002; 62: 4061–4064.
Belkhiri A, Zaika A, Pidkovka N, Knuutila S, Moskaluk C, El-Rifai W . Darpp-32: a novel antiapoptotic gene in upper gastrointestinal carcinomas. Cancer Res 2005; 65: 6583–6592.
Zhu S, Belkhiri A, El-Rifai W . DARPP-32 increases interactions between epidermal growth factor receptor and ERBB3 to promote tumor resistance to gefitinib. Gastroenterology 2011; 141: 1738–48 e2.
Zhu S, Hong J, Tripathi MK, Sehdev V, Belkhiri A, El-Rifai W . Regulation of CXCR4-mediated invasion by DARPP-32 in gastric cancer cells. Mol Cancer Res 2012; 11: 86–94.
Goodison S, Urquidi V, Tarin D . CD44 cell adhesion molecules. Mol Pathol 1999; 52: 189–196.
Pall T, Pink A, Kasak L, Turkina M, Anderson W, Valkna A et al. Soluble CD44 interacts with intermediate filament protein vimentin on endothelial cell surface. PLoS One 2011; 6: e29305.
Hiraga T, Ito S, Nakamura H . Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res 2013; 73: 4112–4122.
Klingbeil P, Natrajan R, Everitt G, Vatcheva R, Marchio C, Palacios J et al. CD44 is overexpressed in basal-like breast cancers but is not a driver of 11p13 amplification. Breast Cancer Res Treat 2010; 120: 95–109.
Screaton GR, Bell MV, Jackson DG, Cornelis FB, Gerth U, Bell JI . Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci USA 1992; 89: 12160–12164.
Omara-Opyene AL, Qiu J, Shah GV, Iczkowski KA . Prostate cancer invasion is influenced more by expression of a CD44 isoform including variant 9 than by Muc18. Lab Invest 2004; 84: 894–907.
Heider KH, Dammrich J, Skroch-Angel P, Muller-Hermelink HK, Vollmers HP, Herrlich P et al. Differential expression of CD44 splice variants in intestinal- and diffuse-type human gastric carcinomas and normal gastric mucosa. Cancer Res 1993; 53: 4197–4203.
Muramaki M, Miyake H, Kamidono S, Hara I . Over expression of CD44V8-10 in human bladder cancer cells decreases their interaction with hyaluronic acid and potentiates their malignant progression. J Urol 2004; 171: 426–430.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456: 470–476.
Xi L, Feber A, Gupta V, Wu M, Bergemann AD, Landreneau RJ et al. Whole genome exon arrays identify differential expression of alternatively spliced, cancer-related genes in lung cancer. Nucleic Acids Res 2008; 36: 6535–6547.
Long JC, Caceres JF . The SR protein family of splicing factors: master regulators of gene expression. Biochem J 2009; 417: 15–27.
Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ et al. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 2012; 19: 96–102.
Cavaloc Y, Bourgeois CF, Kister L, Stevenin J . The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 1999; 5: 468–483.
Matlin AJ, Clark F, Smith CW . Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 2005; 6: 386–398.
Jia R, Li C, McCoy JP, Deng CX, Zheng ZM . SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci 2010; 6: 806–826.
Biamonti G, Ghigna C, Caporali R, Montecucco C . Heterogeneous nuclear ribonucleoproteins (hnRNPs): an emerging family of autoantigens in rheumatic diseases. Clin Exp Rheumatol 1998; 16: 317–326.
Ponta H, Sherman L, Herrlich PA . CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 2003; 4: 33–45.
da Cunha CB, Oliveira C, Wen X, Gomes B, Sousa S, Suriano G et al. De novo expression of CD44 variants in sporadic and hereditary gastric cancer. Lab Invest 2010; 90: 1604–1614.
Younis I, Berg M, Kaida D, Dittmar K, Wang C, Dreyfuss G . Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol 2010; 30: 1718–1728.
Anderson ES, Lin CH, Xiao X, Stoilov P, Burge CB, Black DL . The cardiotonic steroid digitoxin regulates alternative splicing through depletion of the splicing factors SRSF3 and TRA2B. RNA 2012; 18: 1041–1049.
Beckler A, Moskaluk CA, Zaika A, Hampton GM, Powell SM, Frierson HF Jr et al. Overexpression of the 32-kilodalton dopamine and cyclic adenosine 3',5'-monophosphate-regulated phosphoprotein in common adenocarcinomas. Cancer 2003; 98: 1547–1551.
Wang MS, Pan Y, Liu N, Guo C, Hong L, Fan D . Overexpression of DARPP-32 in colorectal adenocarcinoma. Int J Clin Pract 2005; 59: 58–61.
Belkhiri A, Dar AA, Peng DF, Razvi MH, Rinehart C, Arteaga CL et al. Expression of t-DARPP mediates trastuzumab resistance in breast cancer cells. Clin Cancer Res 2008; 14: 4564–4571.
Hamel S, Bouchard A, Ferrario C, Hassan S, Aguilar-Mahecha A, Buchanan M et al. Both t-Darpp and DARPP-32 can cause resistance to trastuzumab in breast cancer cells and are frequently expressed in primary breast cancers. Breast Cancer Res Treat 2009; 120: 47–57.
Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Orntoft TF et al. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J Pathol 2013; 229: 630–639.
Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR . Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol 1997; 138: 225–238.
Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C et al. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol 2009; 83: 167–180.
Miwa T, Watanabe A, Yamada Y, Shino Y, Yamada T, Yamashita J et al. Progression in gastric carcinoma relative to the ratio of CD44 epithelial variant transcript to CD44 hematopoietic variant transcript. Cancer 1996; 77: 25–29.
Prochazka L, Tesarik R, Turanek J . Regulation of alternative splicing of CD44 in cancer. Cell Signal 2014; 26: 2234–2239.
Bourguignon LY, Zhu D, Zhu H . CD44 isoform-cytoskeleton interaction in oncogenic signaling and tumor progression. Front Biosci 1998; 3: d637–d649.
Lau WM, Teng E, Chong HS, Lopez KA, Tay AY, Salto-Tellez M et al. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res 2014; 74: 2630–2641.
Takeuchi K, Yamaguchi A, Urano T, Goi T, Nakagawara G, Shiku H . Expression of CD44 variant exons 8-10 in colorectal cancer and its relationship to metastasis. Jpn J Cancer Res 1995; 86: 292–297.
Pfaffl MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45.
Acknowledgements
This study was supported by grants from the National Institutes of Health (R01CA93999); Vanderbilt SPORE in Gastrointestinal Cancer (P50 CA95103); Vanderbilt Ingram Cancer Center (P30 CA68485); the Vanderbilt Digestive Disease Research Center (DK058404), and the Department of Veterans Affairs. The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health, Department of Veterans Affairs or Vanderbilt University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Zhu, S., Chen, Z., Katsha, A. et al. Regulation of CD44E by DARPP-32-dependent activation of SRp20 splicing factor in gastric tumorigenesis. Oncogene 35, 1847–1856 (2016). https://doi.org/10.1038/onc.2015.250
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.250
This article is cited by
-
Identification of prognostic alternative splicing signature in gastric cancer
Archives of Public Health (2022)
-
A novel SRSF3 inhibitor, SFI003, exerts anticancer activity against colorectal cancer by modulating the SRSF3/DHCR24/ROS axis
Cell Death Discovery (2022)
-
Features of alternative splicing in stomach adenocarcinoma and their clinical implication: a research based on massive sequencing data
BMC Genomics (2020)
-
hnRNPK promotes gastric tumorigenesis through regulating CD44E alternative splicing
Cancer Cell International (2019)
-
Differential networking meta-analysis of gastric cancer across Asian and American racial groups
BMC Systems Biology (2018)