Abstract
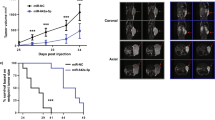
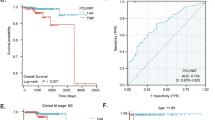
Overwhelming evidence indicates that cancer is a genetic disease caused by the accumulation of mutations in oncogenes and tumor suppressor genes. It is also increasingly apparent, however, that cancer depends not only on mutations in these coding genes but also on alterations in the large class of non-coding RNAs. Here, we report that one such long non-coding RNA, TRPM2-AS, an antisense transcript of TRPM2, which encodes an oxidative stress-activated ion channel, is overexpressed in prostate cancer (PCa). The high expression of TRPM2-AS and its related gene signature were found to be linked to poor clinical outcome, with the related gene signature working also independently of the patient's Gleason score. Mechanistically, TRPM2-AS knockdown led to PCa cell apoptosis, with a transcriptional profile that indicated an unbearable increase in cellular stress in the dying cells, which was coupled to cell cycle arrest, an increase in intracellular hydrogen peroxide and activation of the sense TRPM2 gene. Moreover, targets of existing drugs and treatments were found to be consistently associated with high TRPM2-AS levels in both targeted cells and patients, ultimately suggesting that the measurement of the expression levels of TRPM2-AS allows not only for the early identification of aggressive PCa tumors, but also identifies a subset of at-risk patients who would benefit from currently available, but mostly differently purposed, therapeutic agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Johansson JE, Andrén O, Andersson SO, Dickman PW, Holmberg L, Magnuson A et al. Natural history of early, localized prostate cancer. JAMA 2004; 291: 2713–2719.
Lu-Yao GL, Albertsen PC, Moore DF, Shih W, Lin Y, DiPaola RS et al. Outcomes of localized prostate cancer following conservative management. JAMA 2009; 302: 1202–1209.
Stark JR, Mucci L, Rothman KJ, Adami HO . Screening for prostate cancer remains controversial. BMJ 2009; 339: b3601.
Schröder FH . Landmarks in prostate cancer screening. BJU Int 2012; 1: 3–7.
Xia J, Gulati R, Au M, Gore JL, Lin DW, Etzioni R . Effects of screening on radical prostatectomy efficacy: the prostate cancer intervention versus observation trial. J Natl Cancer Inst 2013; 105: 546–550.
Mattick JS . RNA regulation: a new genetics? Nat Rev Genet 2004; 5: 316–323.
Prasanth KV, Spector DL . Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev 2007; 21: 11–42.
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477: 295–300.
Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013; 493: 231–235.
Gutschner T, Diederichs S . The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012; 9: 703–719.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076.
Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71: 6320–6326.
Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013; 32: 1616–1625.
Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 2011; 29: 742–749.
Orfanelli U, Wenke AK, Doglioni C, Russo V, Bosserhoff AK, Lavorgna G . Identification of novel sense and antisense transcription at the TRPM2 locus in cancer. Cell Res 2008; 18: 1128–1140.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22.
Draghici S, Khatri P, Eklund AC, Szallasi Z . Reliability and reproducibility issues in DNA microarray measurements. Trends Genet 2006; 22: 101–109.
Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 2005; 24: 3847–3852.
Ribeiro F, Paulo P, Costa VL, Barros-Silva JD, Ramalho-Carvalho J, Jerónimo C et al. Cysteine-rich secretory protein-3 (CRISP3) is strongly up-regulated in prostate carcinomas with the TMPRSS2-ERG fusion gene. PLoS ONE 2011; 6: e22317.
Brase JC, Johannes M, Mannsperger H, Fälth M, Metzger J, Kacprzyk LA et al. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-β signaling. BMC Cancer 2011; 11: 507.
Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 2008; 10: 177–188.
Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur SR, Hoshida Y et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics 2010; 16: 3–8.
Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL . Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest 2004; 113: 913–923.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Wang G, Yang ZQ, Zhang K . Endoplasmic reticulum stress response in cancer: molecular mechanism and therapeutic potential. Am J Transl Res 2010; 2: 65–74.
Bai JZ, Lipski J . Differential expression of TRPM2 and TRPV4 channels and their potential role in oxidative stress-induced cell death in organotypic hippocampal culture. Neurotoxicology 2010; 31: 204–214.
Di A, Gao XP, Qian F, Kawamura T, Han J, Hecquet C et al. The redox-sensitive cation channel TRPM2 modulates phagocyte ROS production and inflammation. Nat Immunol 2011; 13: 29–34.
Chen SJ, Zhang W, Tong Q, Conrad K, Hirschler-Laszkiewicz I, Bayerl M et al. Role of TRPM2 in cell proliferation and susceptibility to oxidative stress. Am J Physiol Cell Physiol 2013; 304: C548–C560.
Miller BA . The role of TRP channels in oxidative stress-induced cell death. J Membr Biol 2006; 209: 31–41.
Zhang W, Hirschler-Laszkiewicz I, Tong Q, Conrad K, Sun SC, Penn L et al. TRPM2 is an ion channel that modulates hematopoietic cell death through activation of caspases and PARP cleavage. Am J Physiol Cell Physiol 2006; 29: C1146–C1159.
Kaneko S, Kawakami S, Hara Y, Conrad K, Sun SC, Penn L et al. A critical role of TRPM2 in neuronal cell death by hydrogen peroxide. J Pharmacol Sci 2006; 101: 66–76.
Ishii M, Oyama A, Hagiwara T, Miyazaki A, Mori Y, Kiuchi Y et al. Facilitation of H2O2-induced A172 human glioblastoma cell death by insertion of oxidative stress-sensitive TRPM2 channels. Anticancer Res 2007; 27: 3987–3992.
Yamamoto S, Shimizu S, Kiyonaka S, Takahashi N, Wajima T, Hara Y et al. TRPM2-mediated Ca2+ influx induces chemokine production in monocytes that aggravates inflammatory neutrophil infiltration. Nat Med 2008; 14: 738–747.
Verma S, Quillinan N, Yang YF, Nakayama S, Cheng J, Kelley MH et al. TRPM2 channel activation following in vitro ischemia contributes to male hippocampal cell death. Neurosci Lett 2012; 530: 41–46.
Zeng X, Sikka SC, Huang L, Sun C, Xu C, Jia D et al. Novel role for the transient receptor potential channel TRPM2 in prostate cancer cell proliferation. Prostate Cancer Prostatic Dis 2010; 13: 195–201.
Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G . In search of antisense. Trends Biochem Sci 2004; 29: 88–94.
Cheville JC, Karnes RJ, Therneau TM, Helgeson BE, Cao X, Morris DS et al. Gene panel model predictive of outcome in men at high-risk of systemic progression and death from prostate cancer after radical retropubic prostatectomy. J Clin Oncol 2008; 26: 3930–3936.
Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with PCa: a retrospective study. Lancet Oncol 2011; 12: 245–255.
Penney KL, Sinnott JA, Fall K, Pawitan Y, Hoshida Y, Kraft P et al. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol 2011; 29: 2391–2396.
Markert EK, Mizuno H, Vazquez A, Levine AJ . Molecular classification of prostate cancer using curated expression signatures. Proc Natl Acad Sci USA 2011; 108: 21276–21281.
Agell L, Hernández S, Nonell L, Lorenzo M, Puigdecanet E, de Muga et al. A 12-gene expression signature is associated with aggressive histological in prostate cancer: SEC14L1 and TCEB1 genes are potential markers of progression. Am J Pathol 2012; 18: 1585–1594.
Gasi Tandefelt D, Boormans JL, van der Korput HA, Jenster GW, Trapman J . A 36-gene signature predicts clinical progression in a subgroup of ERG-positive PCas. Eur Urol 2013; 64: 941–950 pii: S0302-2838(13)00222-4.
Irshad S, Bansal M, Castillo-Martin M, Zheng T, Aytes A, Wenske et al. A molecular signature predictive of indolent prostate cancer. Sci Transl Med 2013; 5: 202ra122.
Barabási AL, Gulbahce N, Loscalzo J . Network medicine: a network-based approach to human disease. Nat Rev Genet 2011; 12: 56–68.
US Department of Health and Human Services, FDA, Center for Drug Evaluation and Research & Center for Biologics Evaluation and Research Guidance for industry: clinical trial endpoints for the approval of cancer drugs and biologics (2007).
Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 2011; 470: 269–273.
Hessels D, Schalken JA . The use of PCA3 in the diagnosis of PCa. Nat Rev Urol 2009; 6: 255–261.
Davidson BL, McCray PB. Jr Current prospects for RNA interference-based therapies. Nat Rev Genet 2011; 12: 329–340.
Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 2012; 30: 453–459.
Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM et al. GO::TermFinder–open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 2004; 20: 3710–3715.
Miller EW, Tulyanthan O, Isacoff EY, Chang CJ . Molecular imaging of hydrogen peroxide produced for cell signaling. Nat Chem Biol 2007; 3: 263–267.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B . Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–628.
Xing Y, Kapur K, Wong WH . Probe selection and expression index computation of Affymetrix Exon Arrays. PLoS ONE 2006; 1: e88.
Simon RM, Paik S, Hayes DF . Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009; 101: 1446–1452.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507.
Acknowledgements
This work was supported by grants from the Alleanza Contro il Cancro (AACR) to GL, the Italian Ministry of Health to MB and the Associazione Italiana per la Ricerca sul Cancro (AIRC) to MB.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
This work is the subject of a US provisional patent application.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Orfanelli, U., Jachetti, E., Chiacchiera, F. et al. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene 34, 2094–2102 (2015). https://doi.org/10.1038/onc.2014.144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.144
This article is cited by
-
Long non-coding RNAs in genitourinary malignancies: a whole new world
Nature Reviews Urology (2019)
-
TRPM2 mediates distruption of autophagy machinery and correlates with the grade level in prostate cancer
Journal of Cancer Research and Clinical Oncology (2019)
-
A RNA-Sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer
Scientific Reports (2018)
-
Long non-coding RNA TRPM2-AS as a potential biomarker for hepatocellular carcinoma
Irish Journal of Medical Science (1971 -) (2018)
-
Characters, functions and clinical perspectives of long non-coding RNAs
Molecular Genetics and Genomics (2016)