Abstract
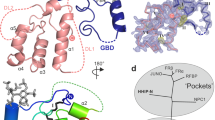
Erythropoetin-producing hepatoma (Eph) receptors are cell-surface protein tyrosine kinases mediating cell-cell communication. Upon activation, they form signaling clusters. We report crystal structures of the full ectodomain of human EphA2 (eEphA2) both alone and in complex with the receptor-binding domain of the ligand ephrinA5 (ephrinA5 RBD). Unliganded eEphA2 forms linear arrays of staggered parallel receptors involving two patches of residues conserved across A-class Ephs. eEphA2–ephrinA5 RBD forms a more elaborate assembly, whose interfaces include the same conserved regions on eEphA2, but rearranged to accommodate ephrinA5 RBD. Cell-surface expression of mutant EphA2s showed that these interfaces are critical for localization at cell-cell contacts and activation-dependent degradation. Our results suggest a 'nucleation' mechanism whereby a limited number of ligand-receptor interactions 'seed' an arrangement of receptors which can propagate into extended signaling arrays.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pasquale, E.B. Eph-ephrin bidirectional signaling in physiology and disease. Cell 133, 38–52 (2008).
Eph Nomenclature Committee. Unified nomenclature for Eph family receptors and their ligands, the ephrins. Cell 90, 403–404 (1997).
Himanen, J.P. & Nikolov, D.B. Eph signaling: a structural view. Trends Neurosci. 26, 46–51 (2003).
Himanen, J.P., Saha, N. & Nikolov, D.B. Cell-cell signaling via Eph receptors and ephrins. Curr. Opin. Cell Biol. 19, 534–542 (2007).
Gale, N.W. et al. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron 17, 9–19 (1996).
Himanen, J.P. et al. Crystal structure of an Eph receptor–ephrin complex. Nature 414, 933–938 (2001).
Cunningham, B.C. et al. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science 254, 821–825 (1991).
Himanen, J.P. et al. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat. Neurosci. 7, 501–509 (2004).
Chrencik, J.E. et al. Structural and biophysical characterization of the EphB4*ephrinB2 protein-protein interaction and receptor specificity. J. Biol. Chem. 281, 28185–28192 (2006).
Himanen, J.P. et al. Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2–ephrin-A1 complex. EMBO Rep. 10, 722–728 (2009).
Bowden, T.A. et al. Structural plasticity of Eph receptor A4 facilitates cross-class ephrin signaling. Structure 17, 1386–1397 (2009).
Smith, F.M. et al. Dissecting the EphA3–Ephrin-A5 interactions using a novel functional mutagenesis screen. J. Biol. Chem. 279, 9522–9531 (2004).
Lindberg, R.A. & Hunter, T. cDNA cloning and characterization of eck, an epithelial cell receptor protein-tyrosine kinase in the eph/elk family of protein kinases. Mol. Cell. Biol. 10, 6316–6324 (1990).
Miao, H., Burnett, E., Kinch, M., Simon, E. & Wang, B. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat. Cell Biol. 2, 62–69 (2000).
Carter, N., Nakamoto, T., Hirai, H. & Hunter, T. EphrinA1-induced cytoskeletal re-organization requires FAK and p130(cas). Nat. Cell Biol. 4, 565–573 (2002).
Lin, Y.G. et al. EphA2 overexpression is associated with angiogenesis in ovarian cancer. Cancer 109, 332–340 (2007).
Zelinski, D.P., Zantek, N.D., Stewart, J.C., Irizarry, A.R. & Kinch, M.S. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 61, 2301–2306 (2001).
Kinch, M.S. & Carles-Kinch, K. Overexpression and functional alterations of the EphA2 tyrosine kinase in cancer. Clin. Exp. Metastasis 20, 59–68 (2003).
Wykosky, J. & Debinski, W. The EphA2 receptor and ephrinA1 ligand in solid tumors: function and therapeutic targeting. Mol. Cancer Res. 6, 1795–1806 (2008).
Reeves, P.J., Callewaert, N., Contreras, R. & Khorana, H.G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK 293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA 99, 13419–13424 (2002).
Aricescu, A.R., Lu, W. & Jones, E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 (2006).
Walter, T.S. et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14, 1617–1622 (2006).
Nikolov, D., Li, C., Lackmann, M., Jeffrey, P. & Himanen, J. Crystal structure of the human ephrin-A5 ectodomain. Protein Sci. 16, 996–1000 (2007).
Cooper, M.A. et al. Loss of ephrin-A5 function disrupts lens fiber cell packing and leads to cataract. Proc. Natl. Acad. Sci. USA 105, 16620–16625 (2008).
Zantek, N.D. et al. E-cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth Differ. 10, 629–638 (1999).
Miura, K., Nam, J.M., Kojima, C., Mochizuki, N. & Sabe, H. EphA2 engages Git1 to suppress Arf6 activity modulating epithelial cell-cell contacts. Mol. Biol. Cell 20, 1949–1959 (2009).
Orsulic, S. & Kemler, R. Expression of Eph receptors and ephrins is differentially regulated by E-cadherin. J. Cell Sci. 113, 1793–1802 (2000).
Shaw, G., Morse, S., Ararat, M. & Graham, F.L. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16, 869–871 (2002).
Carvalho, R.F. et al. Silencing of EphA3 through a cis interaction with ephrinA5. Nat. Neurosci. 9, 322–330 (2006).
Pasquale, E.B. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 6, 462–475 (2005).
Lin, K.T., Sloniowski, S., Ethell, D.W. & Ethell, I.M. Ephrin-B2–induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. J. Biol. Chem. 283, 28969–28979 (2008).
Litterst, C. et al. Ligand binding and calcium influx induce distinct ectodomain/γ-secretase–processing pathways of EphB2 receptor. J. Biol. Chem. 282, 16155–16163 (2007).
Wimmer-Kleikamp, S.H., Janes, P.W., Squire, A., Bastiaens, P.I. & Lackmann, M. Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J. Cell Biol. 164, 661–666 (2004).
Vearing, C.J. & Lackmann, M. Eph receptor signalling; dimerisation just isn't enough. Growth Factors 23, 67–76 (2005).
Egea, J. et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron 47, 515–528 (2005).
Davis, S. et al. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 266, 816–819 (1994).
Bocharov, E.V. et al. Spatial structure and pH-dependent conformational diversity of dimeric transmembrane domain of the receptor tyrosine kinase EphA1. J. Biol. Chem. 283, 29385–29395 (2008).
Thanos, C.D., Goodwill, K.E. & Bowie, J.U. Oligomeric structure of the human EphB2 receptor SAM domain. Science 283, 833–836 (1999).
Diedrichs, K. & Karplus, P.A. Improved R-factors for diffraction data analysis in macromolecular crystallogaphy. Nat. Struct. Biol. 4, 269–275 (1997).
Bendtsen, J.D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Chin, C.N., Sachs, J.N. & Engelman, D.M. Transmembrane homodimerization of receptor-like protein tyrosine phosphatases. FEBS Lett. 579, 3855–3858 (2005).
Grueninger-Leitch, F., D'Arcy, A., D'Arcy, B. & Chene, C. Deglycosylation of proteins for crystallization using recombinant fusion protein glycosidases. Protein Sci. 5, 2617–2622 (1996).
Chang, V.T. et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure 15, 267–273 (2007).
Walter, T.S. et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 61, 651–657 (2005).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Leslie, A.G. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 (2006).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Cowtan, K. An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 34–38 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 (2007).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Collaborative Computational Project 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
DeLano, W.L. The PyMOL Molecular Graphics System. (DeLano Scientific, San Carlos, California, USA, 2002).
Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999).
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005).
Acknowledgements
We thank M. Walsh for assistance during data collection, B. Janssen for aiding with crystallographic data analysis, T.S. Walter for advice on protein crystallization, M. Jones and W. Lu for technical assistance, R. Gilbert for analytical ultracentrifugation measurements, the Molecular Cytogenetics and Microscopy Core facility of the Wellcome Trust Centre for Human Genetics, in particular K.J. Morris and N. Alsamhouri, for their help with confocal microscopy and D.I. Stuart for critical reading of the manuscript. This research was funded by Cancer Research UK. A.R.A. is funded by the UK Medical Research Council and E.Y.J. by Cancer Research UK. The work was supported by the Wellcome Trust Core Award, grant no. 075491/Z/04.
Author information
Authors and Affiliations
Contributions
E.S. conducted crystallographic and cellular studies; K.H. performed crystal mounting and oversaw X-ray data collection; G.S. led the MALS analysis; A.R.A. and E.Y.J. participated in study design and oversaw all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Methods and Supplementary Table 1 (PDF 5250 kb)
Supplementary Video 1
Z-stack images of HEK293T cells expressing EphA2 as mVenus-fusion protein (green). EphA2 localizes at cell-cell contacts. (MOV 853 kb)
Supplementary Video 2
Z-stack images of HEK293T cells expressing the EphA2 interface D mutant L254D+V255D as mVenus fusion protein (green). Mutant EphA2 does not localize at cell-cell contacts. (MOV 654 kb)
Supplementary Video 3
Z-stack images of HEK293T cells expressing the EphA2 deletion construct eEphA2EphA2 as mVenus fusion protein (green). This construct lacks all C-terminal intracellular EphA2 domains. eEphA2EphA2 localizes at cell-cell contacts. (MOV 525 kb)
Supplementary Video 4
Z-stack images of HEK293T cells expressing the EphA2 deletion construct eEphA2Δe as mVenus fusion protein (green). This construct lacks the four N-terminal extracellular EphA2 domains LBD, sushi, EGF, FN1. eEphA2Δe does not localize at cell-cell contacts. (MOV 665 kb)
Rights and permissions
About this article
Cite this article
Seiradake, E., Harlos, K., Sutton, G. et al. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol 17, 398–402 (2010). https://doi.org/10.1038/nsmb.1782
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1782
This article is cited by
-
Eph receptors and ephrins in cancer progression
Nature Reviews Cancer (2024)
-
Research hotspots and trends for axon regeneration (2000–2021): a bibliometric study and systematic review
Inflammation and Regeneration (2022)
-
Cell-cell contact-driven EphB1 cis- and trans- signalings regulate cancer stem cells enrichment after chemotherapy
Cell Death & Disease (2022)
-
Regulation of the EphA2 receptor intracellular region by phosphomimetic negative charges in the kinase-SAM linker
Nature Communications (2021)
-
Aggressive and recurrent ovarian cancers upregulate ephrinA5, a non-canonical effector of EphA2 signaling duality
Scientific Reports (2021)