Abstract
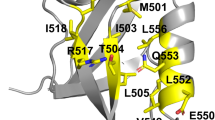
Truncation and mutation of a poorly folded 39-residue peptide has produced 20-residue constructs that are >95% folded in water at physiological pH. These constructs optimize a novel fold, designated as the 'Trp-cage' motif, and are significantly more stable than any other miniprotein reported to date. Folding is cooperative and hydrophobically driven by the encapsulation of a Trp side chain in a sheath of Pro rings. As the smallest protein-like construct, Trp-cage miniproteins should provide a testing ground for both experimental studies and computational simulations of protein folding and unfolding pathways. Pro–Trp interactions may be a particularly effective strategy for the a priori design of self-folding peptides.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Dahiyat, B.I. & Mayo, S.L. Science 278, 82–87 (1997).
Hill, R.B. & DeGrado, W.F. J. Am. Chem. Soc. 120, 1138–1145 (1998).
Walsh, S.T.R., Cheng, H., Bryson, J.W., Roder, H. & DeGrado, W.F. Proc. Natl. Acad. Sci. USA 96, 5486–5491 (1999).
Ottesen, J.J. & Imperiali, B. Nature Struct. Biol. 8, 535–539 (2001).
Cochran, A.G., Skelton, N.J. & Starovasnik, M.A. Proc. Natl. Acad. Sci. USA 98, 5578–5583 (2001).
Li, X., Sutcliffe, M.J., Schwartz, T.W. & Dobson, C.M. Biochemistry 31, 1245–1253 (1992).
Sudol, M. Prog. Biophys. Mol. Biol. 65, 113–132 (1996).
McKnight, C.J., Doering, D.S., Matsudaira, P.T. & Kim, P.S. J. Mol. Biol. 260, 126–134 (1996).
Jager, M., Nguyen, H., Crane, J.C., Kelly, J.W. & Gruebele, M. J. Mol. Biol. 311, 373–393 (2001).
Kortemme, T., Ramírez-Alvarado, M. & Serrano, L. Science 281, 253–256 (1998).
López de la Paz, M., Lacroix, E., Ramírez-Alvarado, M. & Serrano, L. J. Mol. Biol. 312, 229–246 (2001).
Schenck, H. & Gellman, S. J. Am. Chem. Soc. 120, 4869–4870 (1998).
Maynard, A.J., Sharman, G.J. & Searle, M.S. J. Am. Chem. Soc. 120, 1996–2007 (1998).
Andersen, N.H. et al. J. Am. Chem. Soc. 121, 9879–9880 (1999).
Neidigh, J.W., Fesinmeyer, R.M., Prickett, K.S. & Andersen, N.H. Biochemistry 40, 13188–13200 (2001).
Andersen, N.H. & Tong, H. Protein Sci. 6, 1920–1936 (1997).
Huyghues-Despointes, B.M., Klinger, T.M. & Baldwin, R.L. Biochemistry 34, 13267–13271 (1995).
Muñoz, V. & Serrano, L. Biopolymers 41, 495–509 (1997).
Andersen, N.H. et al. J. Am. Chem. Soc. 119, 8547–8561 (1997).
Koradi, R., Billeter, M. & Wüthrich, K. J. Mol. Graph. 14, 51–55 (1996).
Loladze, V.V., Ibarra-Molero, B., Sanxhez-Ruiz, J.M. & Makhatadze, G.I. Biochemistry 38, 16419–16423 (1999).
Andersen, N.H., Cort, J.R., Liu, Z., Sjoberg, S.J. & Tong, H. J. Am. Chem. Soc. 118, 10309–10310 (1996).
Walgers, R., Lee, T.C. & Cammers-Goodwin, A. J. Am. Chem. Soc. 120, 5073–5079 (1998).
Blanco, F.J. & Serrano, L. Eur. J. Biochem. 230, 634–649 (1995).
Ramírez-Alvarado, M., Blanco, F.J. & Serrano, L. Protein Sci. 10, 1381–1392 (2001).
Andersen, N.H., Liu, Z. & Prickett, K.S. FEBS Lett. 399, 47–52 (1996).
Piotto, M., Saudek, V. & Sklenar, V. J. Biomol. NMR 2, 661–665 (1992).
Wüthrich, K. NMR of Proteins and Nucleic Acids (John Wiley, New York; 1986).
Brünger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Acknowledgements
Initial support came from a feasibility grant from the University of Washington Royalty Research Fund with continuing support from an NIH grant. We thank L. Serrano (EMBL-Heidelberg) for reminding us of the pH dependence of the helix-favoring QXXXD interaction.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Neidigh, J., Fesinmeyer, R. & Andersen, N. Designing a 20-residue protein. Nat Struct Mol Biol 9, 425–430 (2002). https://doi.org/10.1038/nsb798
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb798
This article is cited by
-
High resolution ensemble description of metamorphic and intrinsically disordered proteins using an efficient hybrid parallel tempering scheme
Nature Communications (2021)
-
Full structural ensembles of intrinsically disordered proteins from unbiased molecular dynamics simulations
Communications Biology (2021)
-
Sampling of the conformational landscape of small proteins with Monte Carlo methods
Scientific Reports (2020)
-
Hydration shell differentiates folded and disordered states of a Trp-cage miniprotein, allowing characterization of structural heterogeneity by wide-line NMR measurements
Scientific Reports (2019)
-
Designing cooperatively folded abiotic uni- and multimolecular helix bundles
Nature Chemistry (2018)