Abstract
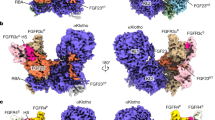
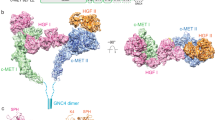
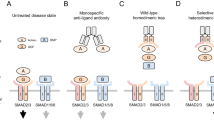
Although ligand-induced receptor dimerization is a common prerequisite for receptor activation, the mode by which different growth factors bind their receptors and cause them to dimerize varies considerably. Here we report the crystal structure at 2.5 Å resolution of NK1, a receptor-binding fragment and a natural splice variant of hepatocyte growth factor/scatter factor (HGF/SF). NK1 assembles as a homodimer in the asymmetric unit, revealing a novel mode of growth factor dimerization produced by close packing of the N domain of one subunit and the kringle domain of the other, thus bringing the two linkers in close proximity. The structure suggests the presence of a binding site for heparan sulfate chains and a mechanism by which the NK1 dimer may engage two receptor molecules through clusters of amino acids located on each protomer and on opposite surfaces of the homodimer. We also report that short (14-mer) heparin fragments effectively dimerize NK1 in solution, implying that heparan sulfate chains may stabilize the NK1 dimer. These results provide a basis for the agonistic activity of NK1 and have implications for the mechanism of receptor binding of HGF/SF.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Donate, L.E. et al. Molecular evolution and domain structure of plasminogen-related growth factors (HGF/SF and HGF1/MSP). Prot. Sci. 3, 2378–2394 (1994).
Bottaro, D.P. et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 251, 802–804 (1991).
Naldini, L. et al. Scatter factor and hepatocyte growth factor are indistinguishable ligands for the MET receptor. EMBO J. 10, 2867–2878 (1991).
Stoker, M., Gherardi, E., Perryman, M. & Gray, J. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature 327, 239–242 (1987).
Gherardi, E., Gray, J., Stoker, M., Perryman, M. & Furlong, R. Purification of scatter factor, a fibroblast-derived basic protein that modulates epithelial interactions and movement. Proc. Natl. Acad. Sci. USA 86, 5844–5848 (1989).
Nakamura, T. et al. Molecular cloning and expression of human hepatocyte growth factor. Nature 342, 440–443 (1989).
Miyazawa, K. et al. Molecular cloning and sequence analysis of cDNA for human hepatocyte growth factor. Biochem. Biophys. Res. Commun. 163, 967–973 (1989).
Rubin, J.S. et al. A broad-spectrum human lung fibroblast-derived mitogen is a variant of hepatocyte growth factor. Proc. Natl. Acad. Sci. USA 88, 415–419 (1991).
Montesano, R., Matsumoto, K., Nakamura, T. & Orci, L. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell 67, 901–908 (1991).
Schmidt, C. et al. Scatter factor/hepatocyte growth factor is essential for liver development. Nature 373, 699–702 (1995).
Uehara, Y. et al. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 373, 702–705 (1995).
Bladt, F., Riethmacher, D., Isenmann, S., Aguzzi, A. & Birchmeier, C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 768–771 (1995).
Gohda, E., Hayashi, Y., Kawaida, A., Tsubouchi, H. & Yamamoto, I. Hepatotrophic growth factor in blood of mice treated with carbon tetrachloride. Life Sci. 46, 1801–1808 (1990).
Nagaike, M. et al. Renotropic functions of hepatocyte growth factor in renal regeneration after unilateral nephrectomy. J. Biol. Chem. 266, 22781–22784 (1991).
Yanagita, K. et al. Hepatocyte growth factor may act as a pulmotrophic factor on lung regeneration after acute lung injury. J. Biol. Chem. 268, 21212–21217 (1993).
Jeffers, M., Rong, S. & Woude, G.F. Hepatocyte growth factor/scatter factor-Met signaling in tumorigenicity and invasion/metastasis. J. Mol. Med. 74, 505–513 (1996).
Lokker, N.A. et al. Structure–function analysis of hepatocyte growth factor. Identification of variants that lack mitogenic activity yet retain high-affinity receptor binding. EMBO J. 11, 2503–2510 (1992).
Okigaki, M., Komada, M., Uehara, Y., Miyazawa, K. & Kitamura, N. Functional characterization of human hepatocyte growth factor mutants obtained by deletion of structural domains. Biochemistry 13, 31, 9555–9561 (1992).
Hartmann, G. et al. A functional domain in the heavy chain of scatter factor/hepatocyte growth factor binds the c-Met receptor and induces cell dissociation but not mitogenesis. Proc. Natl. Acad. Sci. USA 89, 11574–11578 (1992).
Naldini, L. et al. Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO J. 11, 4825–4833 (1992).
Mars, W.M., Zarnegar, R. & Michalopoulos, G.K. Activation of hepatocyte growth factor by the plasminogen activators uPA and tPA. Am. J. Pathol. 143, 949–958 (1993).
Miyazawa, K. et al. Molecular cloning and sequence analysis of the cDNA for a human serine protease responsible for activation of hepatocyte growth factor. Structural similarity of the protease precursor to blood coagulation factor XII. J. Biol. Chem. 268, 10024–10028 (1993).
Matsumoto, K. et al. Deletion of kringle domains or the N-terminal hairpin structure in hepatocyte growth factor results in marked decreases in related biological activities. Biochem. Biophys. Res. Commun. 181, 691–699 (1991).
Hartmann, G. et al. Engineered mutants of HGF/SF with reduced binding to heparan sulfate proteoglycans, decreased clearance and enhanced activity in vivo. Curr. Biol. 8, 125–134 (1998).
Cioce, V. et al. Hepatocyte growth factor (HGF)/NK1 is a naturally occurring HGF/scatter factor variant with partial agonist/antagonist activity. J. Biol. Chem. 271, 13110–13115 (1996).
Miyazawa, K., Kitamura, A., Naka, D. & Kitamura, N. An alternatively processed mRNA generated from human hepatocyte growth factor gene. Eur. J. Biochem. 197, 15–22 (1991).
Chan, A.M. et al. Identification of a competitive HGF antagonist encoded by an alternative transcript. Science 254, 1382–1385 (1991).
Lokker, N.A. & Godowski, P.J. Generation and characterization of a competitive antagonist of human hepatocyte growth factor, HGF/NK1. J. Biol. Chem. 268, 17145–17150 (1993).
Jakubczak, J.L., LaRochelle, W.J. & Merlino, G. NK1, a natural splice variant of hepatocyte growth factor/scatter factor, is a partial agonist in vivo. Mol. Cell. Biol. 18, 1275–1283 (1998).
Takayama, H. et al. Diverse tumorigenesis associated with aberrant development in mice overexpressing hepatocyte growth factor/scatter factor. Proc. Natl. Acad. Sci. USA 94, 701–706 (1997).
Zhou, H. et al. The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site. Structure 6, 109–115 (1998).
Wu, T.P., Padmanabhan, K.P. & Tulinsky, A. The structure of recombinant plasminogen kringle 1 and the fibrin binding site. Blood Coagul. Fibrinolysis 5, 157–166 (1994).
Tickle, I.J., Laskowski, R.A. & Moss, D.S. Rfree and the Rfree Ratio. I. Derivation of expected values of cross-validation residuals used in macromolecular least-squares refinement. Acta Crystallogr. D 54, 547–557 (1998).
Mulichak, A.M., Tulinsky, A. & Ravichandran, K.G. Crystal and molecular structure of human plasminogen kringle 4 refined at 1.9-A resolution. Biochemistry 30, 10576–10588 (1991).
Tulinsky, A., Park, C.H. & Skrzypczak Jankun, E. Structure of prothrombin fragment 1 refined at 2.8 Å resolution. J. Mol. Biol. 202, 885–901 (1988).
Lokker, N.A., Presta, L.G. & Godowski, P.J. Mutational analysis and molecular modeling of the N-terminal kringle-containing domain of hepatocyte growth factor identifies amino acid side chains important for interaction with the c-Met receptor. Protein Engng. 7, 895–903 (1994).
Schwall, R.H. et al. Heparin induces dimerization and confers proliferative activity onto the hepatocyte growth factor antagonists NK1 and NK2. J. Cell Biol. 133, 709–718 (1996).
Mizuno, K. et al. Hairpin loop and second kringle domain are essential sites for heparin binding and biological activity of hepatocyte growth factor. J. Biol. Chem. 269, 1131–1136 (1994).
McDonald, N.Q. et al. New protein fold revealed by a 2.3-Å resolution crystal structure of nerve growth factor. Nature 354, 411–414 (1991).
DiGabriele, A. et al. Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature 393, 812–817 (1998).
de Vos, A.M., Ultsch, M. & Kossiakoff, A.A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255, 306–312 (1992).
Scorer, C.A., Clare, J.J., McCombie, W.R., Romanos, M.A. & Sreekrishna, K. Rapid selection using G418 of high copy number transformants of Pichia pastoris for high-level foreign gene expression. Biotechnology 12, 181–184 (1994).
Mulloy, B., Forster, M.J., Jones, C. & Davies D.B. N.m.r. and molecular-modelling studies of the solution conformation of heparin. Biochem. J. 293: 849–858 (1993).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enz. 276, 301–315 (1996).
Collaborative Computational Project, N. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldegaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. X-PLOR version 3.843: a system for X-ray crystallography and NMR (Yale University Press, New Haven; Connecticut; 1992).
Kleywegt, G.J. & Jones, T.A. Halloween, masks and bones. In From first map to final model (eds Bailey, S., Hubbard, R. & Woller, D.) 59–66 (SERC Daresbury Laboratory, Warrington, UK; 1994).
Evans, S.V. SETOR, hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11, 134–149 (1993).
Nicholls, A., Sharp, K. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Jones, S. & Thornton, J.M. Principles of protein–protein interactions derived from structural studies. Proc. Natl. Acad. Sci. USA 93, 13–20 (1996).
Acknowledgements
We thank S. Batley for assistance with the NK1 constructs and D. Lietha, H. de Jonge and B. Mulloy (National Institute for Biological Standards and Controls) for contributions to the early analysis of NK1–heparin interactions. Work in E.G.'s laboratory was supported by the MRC and the European Union's Biomed Programme. Work in T.L.B.'s laboratory was supported by the Imperial Cancer Research Fund and Wellcome Trust. Work in R.A.B.'s laboratory was supported by the National Cancer Institute, Department of Health and Human Services (DHHS) under contract with ABL. E.G. also has a part time teaching appointment at the Institute C Golgi of the University of Pavia, Italy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chirgadze, D., Hepple, J., Zhou, H. et al. Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding. Nat Struct Mol Biol 6, 72–79 (1999). https://doi.org/10.1038/4947
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/4947
This article is cited by
-
Overexpression-mediated activation of MET in the Golgi promotes HER3/ERBB3 phosphorylation
Oncogene (2019)
-
Macrocyclic peptide-based inhibition and imaging of hepatocyte growth factor
Nature Chemical Biology (2019)
-
H1/pHGFK1 nanoparticles exert anti-tumoural and radiosensitising effects by inhibition of MET in glioblastoma
British Journal of Cancer (2018)
-
Targeting MET in cancer: rationale and progress
Nature Reviews Cancer (2012)