Key Points
-
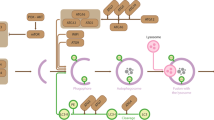
SUMOylation is a reversible covalent post-translational modification that involves the conjugation of a member of the small ubiquitin-like modifier (SUMO) family to lysine residues in target proteins. SUMO proteins are conjugated by an enzymatic pathway and can mediate a diverse range of subsequent fates for substrate proteins.
-
In a number of systems, both covalent attachment of SUMO and non-covalent binding to SUMO contribute to functional outcomes. Recently, a number of SUMO interacting motifs (SIMs) have been reported that allow proteins to bind to SUMO.
-
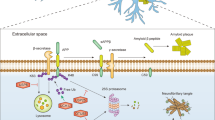
SUMOylation is best characterized in the nucleus; however, numerous extranuclear substrates have also been reported. Potential roles for extranuclear SUMOylation include the regulation of G-protein, kinase and phosphatase signalling, axonal mRNA trafficking, and mitochondrial fission and apoptosis.
-
At the plasma membrane, SUMOylation has been implicated in the regulation of a number of key proteins, including glucose transporters, potassium channels and glutamate receptors.
-
SUMOylation has a central role in synapse formation through the modification of the transcription factor MEF2A. Active MEF2A inhibits synapse formation, and the activity-dependent interplay between SUMOylation, phosphorylation and acetylation of MEF2A can suppress this inhibition.
-
A role for SUMO in various neurological disorders is becoming increasingly well documented. Numerous forms of neuronal inclusions stain positive for SUMO family members, and proteins involved in the pathogenesis of disorders such as Huntington's disease, Parkinson's disease and Alzheimer's disease have been reported to be SUMO substrates.
-
SUMO features in various cellular stress responses. For example, large changes in protein SUMOylation have been reported upon cellular oxidative stress and ischemia.
Abstract
Post-translational protein modifications are integral components of signalling cascades that enable cells to efficiently, rapidly and reversibly respond to extracellular stimuli. These modifications have crucial roles in the CNS, where the communication between neurons is particularly complex. SUMOylation is a post-translational modification in which a member of the small ubiquitin-like modifier (SUMO) family of proteins is conjugated to lysine residues in target proteins. It is well established that SUMOylation controls many aspects of nuclear function, but it is now clear that it is also a key determinant in many extranuclear neuronal processes, and it has also been implicated in a wide range of neuropathological conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Matunis, M. J., Coutavas, E. & Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470 (1996).
Mahajan, R., Delphin, C., Guan, T., Gerace, L. & Melchior, F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107 (1997). Together with reference 1, this was the first study to identify SUMO1 as a covalent modifier protein. Both papers describe the role of SUMO in regulating the nuclear-pore localization of RanGAP.
Hayashi, T. et al. Ubc9 is essential for viability of higher eukaryotic cells. Exp. Cell Res. 280, 212–221 (2002).
Johnson, E. S. & Blobel, G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272, 26799–26802 (1997). This study, along with reference 137, indicated that the post-translational modification of proteins by SUMO in yeast and mammalian cells is mediated by UBC9, which is SUMO-specific and does not conjugate ubiquitin.
Tanaka, K. et al. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell Biol. 19, 8660–8672 (1999).
Lapenta, V. et al. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics 40, 362–366 (1997).
Bohren, K. M., Nadkarni, V., Song, J. H., Gabbay, K. H. & Owerbach, D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279, 27233–27238 (2004).
Johnson, E. S. Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 (2004).
Bayer, P. et al. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275–286 (1998).
Bohren, K. M., Gabbay, K. H. & Owerbach, D. Affinity chromatography of native SUMO proteins using His-tagged recombinant UBC9 bound to Co2+-charged talon resin. Protein Expr. Purif. 54, 289–294 (2007).
Owerbach, D., McKay, E. M., Yeh, E. T., Gabbay, K. H. & Bohren, K. M. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337, 517–520 (2005).
Tatham, M. H. et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 (2001).
Saitoh, H. & Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252–6258 (2000).
Hardeland, U., Steinacher, R., Jiricny, J. & Schar, P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. Embo J. 21, 1456–1464 (2002).
Hofmann, H., Floss, S. & Stamminger, T. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74, 2510–2524 (2000).
Rodriguez, M. S., Dargemont, C. & Hay, R. T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 (2001).
Sampson, D. A., Wang, M. & Matunis, M. J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664–21669 (2001). This study, along with reference 16, defined the SUMOylation consensus motif and demonstrated that direct binding of UBC9 to this motif is required for SUMO modification.
Gong, L., Li, B., Millas, S. & Yeh, E. T. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 448, 185–189 (1999).
Hay, R. T. SUMO: a history of modification. Mol. Cell 18, 1–12 (2005).
Hoeller, D. et al. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell 26, 891–898 (2007).
Bernier-Villamor, V., Sampson, D. A., Matunis, M. J. & Lima, C. D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 (2002).
Johnson, E. S. & Gupta, A. A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 (2001). This was the first report to indicate that E3 enzymes exist for the SUMO pathway.
Takahashi, Y., Kahyo, T., Toh, E. A., Yasuda, H. & Kikuchi, Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276, 48973–48977 (2001).
Takahashi, Y., Toh-e, A. & Kikuchi, Y. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275, 223–231 (2001).
Kahyo, T., Nishida, T. & Yasuda, H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718 (2001).
Schmidt, D. & Muller, S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl Acad. Sci. USA 99, 2872–2877 (2002).
Pichler, A., Gast, A., Seeler, J. S., Dejean, A. & Melchior, F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 (2002).
Kagey, M. H., Melhuish, T. A. & Wotton, D. The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137 (2003).
Mukhopadhyay, D. & Dasso, M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286–295 (2007).
Bossis, G. & Melchior, F. SUMO: regulating the regulator. Cell Div. 1, 13 (2006).
Bencsath, K. P., Podgorski, M. S., Pagala, V. R., Slaughter, C. A. & Schulman, B. A. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 277, 47938–47945 (2002).
Bylebyl, G. R., Belichenko, I. & Johnson, E. S. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278, 44113–44120 (2003).
Fu, C. et al. Stabilization of PML nuclear localization by conjugation and oligomerization of SUMO-3. Oncogene 24, 5401–5413 (2005).
Pedrioli, P. G. et al. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nature Methods 3, 533–539 (2006).
Pichler, A. & Melchior, F. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3, 381–387 (2002).
Yang, M., Hsu, C. T., Ting, C. Y., Liu, L. F. & Hwang, J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J. Biol. Chem. 281, 8264–8274 (2006).
Kerscher, O. SUMO junction—what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 (2007).
Scheschonka, A., Tang, Z. & Betz, H. Sumoylation in neurons: nuclear and synaptic roles? Trends Neurosci. 30, 85–91 (2007).
Mishra, R. K. et al. Dynamin interacts with members of the sumoylation machinery. J. Biol. Chem. 279, 31445–31454 (2004).
Um, J. W. & Chung, K. C. Functional modulation of parkin through physical interaction with SUMO-1. J. Neurosci. Res. 84, 1543–1554 (2006).
Hicke, L. & Dunn, R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 (2003).
Seeler, J. S. & Dejean, A. Nuclear and unclear functions of SUMO. Nature Rev. Mol. Cell Biol. 4, 690–699 (2003).
Heun, P. SUMOrganization of the nucleus. Curr. Opin. Cell Biol. 27 Apr 2007 (doi:10.1016/j.ceb.2007.04.014).
Dohmen, R. J. SUMO protein modification. Biochim. Biophys. Acta 1695, 113–131 (2004).
Melchior, F., Schergaut, M. & Pichler, A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28, 612–618 (2003).
Vassilatis, D. K. et al. The G protein-coupled receptor repertoires of human and mouse. Proc. Natl Acad. Sci. USA 100, 4903–4908 (2003).
Gainetdinov, R. R., Premont, R. T., Bohn, L. M., Lefkowitz, R. J. & Caron, M. G. Desensitization of G protein-coupled receptors and neuronal functions. Annu. Rev. Neurosci. 27, 107–144 (2004).
Bauer, P. H. et al. Phosducin is a protein kinase A-regulated G-protein regulator. Nature 358, 73–76 (1992).
Berman, D. M. & Gilman, A. G. Mammalian RGS proteins: barbarians at the gate. J. Biol. Chem. 273, 1269–1272 (1998).
Mao, H. et al. RGS17/RGSZ2, a novel regulator of Gi/o, Gz, and Gq signaling. J. Biol. Chem. 279, 26314–26322 (2004).
Garzon, J., Rodriguez-Munoz, M., Lopez-Fando, A. & Sanchez-Blazquez, P. The RGSZ2 protein exists in a complex with μ-opioid receptors and regulates the desensitizing capacity of Gz proteins. Neuropsychopharmacology 30, 1632–1648 (2005).
Rodriguez-Munoz, M., Bermudez, D., Sanchez-Blazquez, P. & Garzon, J. Sumoylated RGS-Rz proteins act as scaffolds for Mu-opioid receptors and G-protein complexes in mouse brain. Neuropsychopharmacology 32, 842–850 (2007).
Danner, S. & Lohse, M. J. Phosducin is a ubiquitous G-protein regulator. Proc. Natl Acad. Sci. USA 93, 10145–10150 (1996).
Klenk, C., Humrich, J., Quitterer, U. & Lohse, M. J. SUMO-1 controls the protein stability and the biological function of phosducin. J. Biol. Chem. 281, 8357–8364 (2006).
Robles, E. & Gomez, T. M. Focal adhesion kinase signaling at sites of integrin-mediated adhesion controls axon pathfinding. Nature Neurosci. 9, 1274–1283 (2006).
Kadare, G. et al. PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J. Biol. Chem. 278, 47434–47440 (2003).
Mitra, S. K., Hanson, D. A. & Schlaepfer, D. D. Focal adhesion kinase: in command and control of cell motility. Nature Rev. Mol. Cell Biol. 6, 56–68 (2005).
Dube, N. & Tremblay, M. L. Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: from diabetes, obesity to cell cycle, and cancer. Biochim. Biophys. Acta 1754, 108–117 (2005).
Zabolotny, J. M. et al. PTP1B regulates leptin signal transduction in vivo. Dev. Cell 2, 489–495 (2002).
Cheng, A. et al. Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Dev. Cell 2, 497–503 (2002).
Bence, K. K. et al. Neuronal PTP1B regulates body weight, adiposity and leptin action. Nature Med. 12, 917–924 (2006).
Ahima, R. S. & Flier, J. S. Leptin. Annu. Rev. Physiol. 62, 413–437 (2000).
Dadke, S. et al. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nature Cell Biol. 9, 80–85 (2007).
Rui, H. L. et al. SUMO-1 modification of the C-terminal KVEKVD of Axin is required for JNK activation but has no effect on Wnt signaling. J. Biol. Chem. 277, 42981–42986 (2002).
Martin, K. C. Local protein synthesis during axon guidance and synaptic plasticity. Curr. Opin. Neurobiol. 14, 305–310 (2004).
Giuditta, A., Kaplan, B. B., van Minnen, J., Alvarez, J. & Koenig, E. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci. 25, 400–404 (2002).
Wolin, S. L. & Cedervall, T. The La protein. Annu. Rev. Biochem. 71, 375–403 (2002).
van Niekerk, E. A. et al. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc. Natl Acad. Sci. USA 104, 12913–12918 (2007).
Hirokawa, N. & Takemura, R. Molecular motors and mechanisms of directional transport in neurons. Nature Rev. Neurosci. 6, 201–214 (2005).
Li, Z., Okamoto, K., Hayashi, Y. & Sheng, M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119, 873–887 (2004).
Smirnova, E., Griparic, L., Shurland, D. L. & van der Bliek, A. M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 (2001).
Harder, Z., Zunino, R. & McBride, H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr. Biol. 14, 340–345 (2004).
Wasiak, S., Zunino, R. & McBride, H. M. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 177, 439–450 (2007).
Hayashi, N., Shirakura, H., Uehara, T. & Nomura, Y. Relationship between SUMO-1 modification of caspase-7 and its nuclear localization in human neuronal cells. Neurosci. Lett. 397, 5–9 (2006).
Besnault-Mascard, L. et al. Caspase-8 sumoylation is associated with nuclear localization. Oncogene 24, 3268–3273 (2005).
Zunino, R., Schauss, A., Rippstein, P., Andrade-Navarro, M. & McBride, H. M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 120, 1178–1188 (2007).
Martin, S., Nishimune, A., Mellor, J. R. & Henley, J. M. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature 447, 321–325 (2007). This was the first demonstration that SUMOylation occurs at synapses. This study showed that SUMO modification is involved in agonist-dependent kainate-receptor endocytosis and the modulation of synaptic transmission.
Giorgino, F. et al. The sentrin-conjugating enzyme mUbc9 interacts with GLUT4 and GLUT1 glucose transporters and regulates transporter levels in skeletal muscle cells. Proc. Natl Acad. Sci. USA 97, 1125–1130 (2000). This paper was the first to show the SUMO modification of a plasma membrane protein.
Leloup, C. et al. Discrete brain areas express the insulin-responsive glucose transporter GLUT4. Brain Res. Mol. Brain Res. 38, 45–53 (1996).
Kobayashi, M., Nikami, H., Morimatsu, M. & Saito, M. Expression and localization of insulin-regulatable glucose transporter (GLUT4) in rat brain. Neurosci. Lett. 213, 103–106 (1996).
Liu, L. B., Omata, W., Kojima, I. & Shibata, H. The SUMO conjugating enzyme Ubc9 is a regulator of GLUT4 turnover and targeting to the insulin-responsive storage compartment in 3T3-L1 adipocytes. Diabetes 56, 1977–1985 (2007).
Plant, L. D., Rajan, S. & Goldstein, S. A. K2P channels and their protein partners. Curr. Opin. Neurobiol. 15, 326–333 (2005).
Rajan, S., Plant, L. D., Rabin, M. L., Butler, M. H. & Goldstein, S. A. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell 121, 37–47 (2005).
Feliciangeli, S. et al. Does sumoylation control K2P1/TWIK1 background K+ channels? Cell 130, 563–569 (2007).
Pannasch, U. et al. The potassium channels Kv1.5 and Kv1.3 modulate distinct functions of microglia. Mol. Cell Neurosci. 33, 401–411 (2006).
Benson, M. D. et al. SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc. Natl Acad. Sci. USA 104, 1805–1810 (2007).
Grosse, G. et al. Expression of Kv1 potassium channels in mouse hippocampal primary cultures: development and activity-dependent regulation. J. Neurosci. 20, 1869–1882 (2000).
Tsaur, M. L., Sheng, M., Lowenstein, D. H., Jan, Y. N. & Jan, L. Y. Differential expression of K+ channel mRNAs in the rat brain and down-regulation in the hippocampus following seizures. Neuron 8, 1055–1067 (1992).
Ferraguti, F. & Shigemoto, R. Metabotropic glutamate receptors. Cell Tissue Res. 326, 483–504 (2006).
Tang, Z., El Far, O., Betz, H. & Scheschonka, A. Pias1 interaction and sumoylation of metabotropic glutamate receptor 8. J. Biol. Chem. 280, 38153–38159 (2005).
Enz, R. The trick of the tail: protein-protein interactions of metabotropic glutamate receptors. Bioessays 29, 60–73 (2007).
Pinheiro, P. & Mulle, C. Kainate receptors. Cell Tissue Res. 326, 457–482 (2006).
Isaac, J. T., Mellor, J., Hurtado, D. & Roche, K. W. Kainate receptor trafficking: physiological roles and molecular mechanisms. Pharmacol. Ther. 104, 163–172 (2004).
Jaskolski, F., Coussen, F. & Mulle, C. Subcellular localization and trafficking of kainate receptors. Trends Pharmacol. Sci. 26, 20–26 (2005).
Lerma, J. Roles and rules of kainate receptors in synaptic transmission. Nature Rev. Neurosci. 4, 481–495 (2003).
Shalizi, A. K. & Bonni, A. Brawn for brains: the role of MEF2 proteins in the developing nervous system. Curr. Top. Dev. Biol. 69, 239–266 (2005).
Shalizi, A. et al. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J. Neurosci. 27, 10037–10046 (2007).
Flavell, S. W. et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 (2006).
Shalizi, A. et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311, 1012–1017 (2006). This paper, together with the work in reference 98, demonstrated that MEF2 transcription factors can be modified by phosphorylation, acetylation and SUMOylation. The balance between these modifications acts to control activity-dependent synapse formation.
Weissmann, C. & Brandt, R. Mechanisms of neurodegenerative diseases: insights from live cell imaging. J. Neurosci. Res. 1 Aug 2007 (doi:10.1002/jnr.21448).
Dorval, V. & Fraser, P. E. SUMO on the road to neurodegeneration. Biochim. Biophys. Acta 1773, 694–706 (2007).
Lieberman, A. P., Robitaille, Y., Trojanowski, J. Q., Dickson, D. W. & Fischbeck, K. H. Polyglutamine-containing aggregates in neuronal intranuclear inclusion disease. Lancet 351, 884 (1998).
Pountney, D. L. et al. SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp. Neurol. 184, 436–446 (2003).
Takahashi-Fujigasaki, J., Arai, K., Funata, N. & Fujigasaki, H. SUMOylation substrates in neuronal intranuclear inclusion disease. Neuropathol. Appl. Neurobiol. 32, 92–100 (2006).
McFadden, K. et al. Neuronal intranuclear inclusion disease without polyglutamine inclusions in a child. J. Neuropathol. Exp. Neurol. 64, 545–552 (2005).
Walker, F. O. Huntington's disease. Lancet 369, 218–228 (2007).
Steffan, J. S. et al. SUMO modification of huntingtin and Huntington's disease pathology. Science 304, 100–104 (2004). This was the first study to demonstrate a mechanistic role for SUMOylation in Huntington's disease pathology.
Chan, H. Y., Warrick, J. M., Andriola, I., Merry, D. & Bonini, N. M. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum. Mol. Genet. 11, 2895–2904 (2002).
Ueda, H. et al. Enhanced SUMOylation in polyglutamine diseases. Biochem. Biophys. Res. Commun. 293, 307–313 (2002).
Terashima, T., Kawai, H., Fujitani, M., Maeda, K. & Yasuda, H. SUMO-1 co-localized with mutant atrophin-1 with expanded polyglutamines accelerates intranuclear aggregation and cell death. Neuroreport 13, 2359–2364 (2002).
Zoghbi, H. Y. & Orr, H. T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217–247 (2000).
Dawson, T. M. & Dawson, V. L. Molecular pathways of neurodegeneration in Parkinson's disease. Science 302, 819–822 (2003).
Dorval, V. & Fraser, P. E. Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and α-synuclein. J. Biol. Chem. 281, 9919–9924 (2006).
Pountney, D. L., Chegini, F., Shen, X., Blumbergs, P. C. & Gai, W. P. SUMO-1 marks subdomains within glial cytoplasmic inclusions of multiple system atrophy. Neurosci. Lett. 381, 74–79 (2005).
Um, J. W. et al. Parkin ubiquitinates and promotes the degradation of RanBP2. J. Biol. Chem. 281, 3595–3603 (2006).
Shinbo, Y. et al. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 13, 96–108 (2006).
Suh, Y. H. & Checler, F. Amyloid precursor protein, presenilins, and α-synuclein: molecular pathogenesis and pharmacological applications in Alzheimer's disease. Pharmacol. Rev. 54, 469–525 (2002).
Li, Y. et al. Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc. Natl Acad. Sci. USA 100, 259–264 (2003).
Dorval, V., Mazzella, M. J., Mathews, P. M., Hay, R. T. & Fraser, P. E. Modulation of Aβ generation by small ubiquitin-like modifiers does not require conjugation to target proteins. Biochem. J. 404, 309–316 (2007).
Bossis, G. & Melchior, F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21, 349–357 (2006). This paper demonstrates that ROS are key regulators of the SUMOylation–de-SUMOylation equilibrium.
Lee, Y. J. & Hallenbeck, J. M. Insights into cytoprotection from ground squirrel hibernation, a natural model of tolerance to profound brain oligaemia. Biochem. Soc. Trans. 34, 1295–1298 (2006).
Yang, W., Sheng, H., Warner, D. S. & Paschen, W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J. Cereb. Blood Flow Metab. 13 Jun 2007 (doi:10.1038/sj.jcbfm.9600523).
Cimarosti, H., Lindberg, C., Bomholt, S. F., Rønn, L. C. B. & Henley, J. M. Increased protein SUMOylation following focal cerebral ischemia. Neuropharmacology 6 Oct 2007 (doi:10.1016/j.neuropharm.2007.09.010).
Hurley, J. H., Lee, S. & Prag, G. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006).
Hecker, C. M., Rabiller, M., Haglund, K., Bayer, P. & Dikic, I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 (2006).
Song, J., Zhang, Z., Hu, W. & Chen, Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 (2005).
Minty, A., Dumont, X., Kaghad, M. & Caput, D. Covalent modification of p73α by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275, 36316–36323 (2000).
Song, J., Durrin, L. K., Wilkinson, T. A., Krontiris, T. G. & Chen, Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl Acad. Sci. USA 101, 14373–14378 (2004).
Hannich, J. T. et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280, 4102–4110 (2005).
Yang, S. H., Galanis, A., Witty, J. & Sharrocks, A. D. An extended consensus motif enhances the specificity of substrate modification by SUMO. Embo J. 25, 5083–5093 (2006).
Hietakangas, V. et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl Acad. Sci. USA 103, 45–50 (2006).
Poukka, H., Karvonen, U., Janne, O. A. & Palvimo, J. J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl Acad. Sci. USA 97, 14145–14150 (2000).
Stankovic-Valentin, N. et al. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved ΨKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell Biol. 27, 2661–2675 (2007).
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002).
Johnson, E. S., Schwienhorst, I., Dohmen, R. J. & Blobel, G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. Embo J. 16, 5509–5519 (1997). This was the first study to indicate that SUMO proteins are activated by a heterodimer of AOS1 and UBA2.
Okuma, T., Honda, R., Ichikawa, G., Tsumagari, N. & Yasuda, H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Commun. 254, 693–698 (1999).
Desterro, J. M., Thomson, J. & Hay, R. T. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417, 297–300 (1997).
Kotaja, N., Karvonen, U., Janne, O. A. & Palvimo, J. J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell Biol. 22, 5222–5234 (2002).
Nishida, T. & Yasuda, H. PIAS1 and PIASxα function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 277, 41311–41317 (2002).
Sachdev, S. et al. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15, 3088–3103 (2001).
Zhao, X. & Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. USA 102, 4777–4782 (2005).
Weger, S., Hammer, E. & Heilbronn, R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 579, 5007–5012 (2005).
Morita, Y., Kanei-Ishii, C., Nomura, T. & Ishii, S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol. Biol. Cell 16, 5433–5444 (2005).
Li, S. J. & Hochstrasser, M. A new protease required for cell-cycle progression in yeast. Nature 398, 246–251 (1999).
Li, S. J. & Hochstrasser, M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell Biol. 20, 2367–2377 (2000).
Gong, L., Millas, S., Maul, G. G. & Yeh, E. T. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275, 3355–3359 (2000).
Yeh, E. T., Gong, L. & Kamitani, T. Ubiquitin-like proteins: new wines in new bottles. Gene 248, 1–14 (2000).
Nishida, T., Tanaka, H. & Yasuda, H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267, 6423–6427 (2000).
Kim, K. I. et al. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275, 14102–14106 (2000).
Bossis, G. et al. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol. Cell Biol. 25, 6964–6979 (2005).
Desterro, J. M., Rodriguez, M. S. & Hay, R. T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2, 233–239 (1998).
Ulrich, H. D. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 15, 525–532 (2005).
Sapetschnig, A. et al. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. Embo J. 21, 5206–5215 (2002).
Carter, S., Bischof, O., Dejean, A. & Vousden, K. H. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nature Cell Biol. 9, 428–435 (2007).
Acknowledgements
We are grateful to the MRC, the Wellcome Trust and the EU (GRIPPANT; PL 005320) for financial support.
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Ubiquitylation
-
The covalent attachment of the 76-amino-acid protein ubiquitin to lysine residues in target proteins. Ubiquitylation is typically involved in the sorting of proteins for degradation, although there are numerous exceptions.
- Paralogue
-
Either of a pair of genes that derive from the same ancestral gene.
- Consensus motif
-
A sequence that is conserved among proteins that undergo a particular modification within that sequence. Consensus motifs also exist for several non-covalent interactions.
- Isopeptidase
-
An enzyme that specifically recognizes isopeptide bonds and cleaves them. SENPs have SUMO-specific isopeptidase activity, and cleave SUMO from SUMO-conjugated substrates.
- Deconjugation
-
The removal of covalently attached SUMO from the substrate.
- G protein
-
A heterotrimeric GTP-binding protein that interacts with cell-surface receptors, often stimulating or inhibiting the activity of a downstream enzyme. G proteins consist of three subunits: the α subunit, which contains the guanine-nucleotide-binding site, and the β and γ subunits, which function as a heterodimer.
- Synaptosomes
-
Discrete structures formed from the synaptic terminals upon brain homogenization in which the main structural presynaptic features are preserved. These structures retain the ability take up, store and release neurotransmitters.
- Nucleocytoplasmic shuttling
-
Bidirectional protein transport between the cytoplasm and the nuclear matrix through the nuclear pore complex.
- Synaptic plasticity
-
A cellular process that results in lasting changes in the efficacy of neurotransmission.
- Yeast two-hybrid screen
-
A system used to determine whether proteins directly interact. It involves the use of plasmids that encode two hybrid proteins, one of which is fused to the GAL4 DNA-binding domain and the other of which is fused to the GAL4 activation domain. The two proteins are expressed together in yeast and, if they interact, the resulting complex will drive the expression of a reporter gene, commonly β-galactosidase.
- Metabotropic glutamate receptors
-
A family of eight GPCRs that are activated by glutamate. They are classified into three groups (I–III) on the basis of their pharmacological properties and their downstream effector cascades.
- Dendritic claw
-
A postsynaptic differentiation of dendrites that is morphologically characteristic. In the cerebellum, granule cells develop dendritic claws with which they form synapses with mossy-fibre terminals.
- α-synuclein
-
A neuronal protein of unknown function that is detected mainly in presynaptic terminals. It can aggregate to form insoluble fibrils known as Lewy bodies, which are observed in pathological conditions such as Parkinson's disease.
- Proteasome
-
The protein complex that is responsible for degrading intracellular proteins that have been tagged for destruction by the conjugation of ubiquitin.
- Oxidative stress
-
A disturbance in the oxidant–antioxidant balance in favour of the former, leading to potential cellular damage including the mutation of DNA bases, protein oxidation and the generation of lipid peroxidation products.
- Amyloid-β (Aβ)
-
A peptide of size 39–43 amino acids that is the main constituent of amyloid plaques in the brain of Alzheimer's disease patients. These plaques are composed of a tangle of regularly ordered fibrillar aggregates called amyloid fibres. Among these heterogeneous peptide molecules, Aβ40 and Aβ42 are the most common isoforms. Aβ42 is the most fibrillogenic peptide and is thus associated with disease states.
- Reactive oxygen species
-
(ROS). These include free radicals, peroxides and oxygen ions. ROS form as a natural byproduct of oxidative phosphorylation in the mitochondria. Under pathological conditions, ROS levels can increase significantly, resulting in serious cellular damage. Cells normally defend themselves against ROS damage by the action of enzymes such as catalases and superoxide dismutase.
- Excitotoxicity
-
A pathological process by which neurons are damaged and killed by the overactivation of glutamate receptors.
- Long-term potentiation
-
(LTP). A form of synaptic plasticity that results in a long-lasting increase in the strength of synaptic transmission.
- Long-term depression
-
(LTD). A form of synaptic plasticity that results in a longlasting decrease in the strength of synaptic transmission.
Rights and permissions
About this article
Cite this article
Martin, S., Wilkinson, K., Nishimune, A. et al. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci 8, 948–959 (2007). https://doi.org/10.1038/nrn2276
Issue Date:
DOI: https://doi.org/10.1038/nrn2276
This article is cited by
-
RNF166 plays a dual role for Lys63-linked ubiquitination and sumoylation of its target proteins
Journal of Neural Transmission (2022)
-
DNMT3b SUMOylation Mediated MMP-2 Upregulation Contribute to Paclitaxel Induced Neuropathic Pain
Neurochemical Research (2021)
-
Guanosine modulates SUMO2/3-ylation in neurons and astrocytes via adenosine receptors
Purinergic Signalling (2020)
-
Yy1 regulates Senp1 contributing to AMPA receptor GluR1 expression following neuronal depolarization
Journal of Biomedical Science (2019)
-
Sumoylated α-synuclein translocates into the nucleus by karyopherin α6
Molecular & Cellular Toxicology (2019)