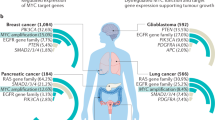
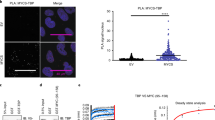
Abstract
MYC is a potent oncogene that drives unrestrained cell growth and proliferation. Shortly after its discovery as an oncogene, the MYC protein was recognized as a sequence-specific transcription factor. Since that time, MYC oncogene research has focused on the mechanism of MYC-induced transcription and on the identification of MYC transcriptional target genes. Recently, MYC was shown to control protein expression through mRNA translation and to directly regulate DNA replication, thus initiating exciting new areas of oncogene research.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Oster, S. K., Ho, C. S., Soucie, E. L. & Penn, L. Z. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 84, 81–154 (2002).
Pirity, M., Blanck, J. K. & Schreiber-Agus, N. Lessons learned from Myc/Max/Mad knockout mice. Curr. Top. Microbiol. Immunol. 302, 205–234 (2006).
Evan, G. I. & Vousden, K. H. Proliferation, cell cycle and apoptosis in cancer. Nature 411, 342–348 (2001).
Dang, C. V. et al. The c-Myc target gene network. Semin. Cancer Biol. 16, 253–264 (2006).
Felsher, D. W. & Bishop, J. M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4, 199–207 (1999).
Vita, M. & Henriksson, M. The Myc oncoprotein as a therapeutic target for human cancer. Semin. Cancer Biol. 16, 318–330 (2006).
Patel, J. H., Loboda, A. P., Showe, M. K., Showe, L. C. & McMahon, S. B. Analysis of genomic targets reveals complex functions of MYC. Nature Rev. Cancer 4, 562–568 (2004).
Adhikary, S. & Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nature Rev. Mol. Cell Biol. 6, 635–645 (2005).
Cowling, V. H. & Cole, M. D. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 16, 242–252 (2006).
Cowling, V. H. & Cole, M. D. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol. Cell. Biol. 27, 2059–2073 (2007).
Dominguez-Sola, D. et al. Non-transcriptional control of DNA replication by c-Myc. Nature 448, 445–451 (2007).
Hurlin, P. J. & Huang, J. The MAX-interacting transcription factor network. Semin. Cancer Biol. 16, 265–274 (2006).
Gomez-Roman, N., Grandori, C., Eisenman, R. N. & White, R. J. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421, 290–294 (2003).
Kenneth, N. S. et al. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Natl Acad. Sci. USA 104, 14917–14922 (2007).
Grandori, C. et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature Cell Biol. 7, 311–318 (2005).
Grewal, S. S., Li, L., Orian, A., Eisenman, R. N. & Edgar, B. A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nature Cell Biol. 7, 295–302 (2005).
Arabi, A. et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nature Cell Biol. 7, 303–310 (2005).
O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 (2005).
Chang, T. C. et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genet. 40, 43–50 (2008).
McMahon, S. B., Van Buskirk, H. A., Dugan, K. A., Copeland, T. D. & Cole, M. D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 (1998).
McMahon, S. B., Wood, M. A. & Cole, M. D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20, 556–562 (2000).
Frank, S. R., Schroeder, M., Fernandez, P., Taubert, S. & Amati, B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15, 2069–2082 (2001).
Vervoorts, J. et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 4, 484–490 (2003).
Lee, K. K. & Workman, J. L. Histone acetyltransferase complexes: one size doesn't fit all. Nature Rev. Mol. Cell Biol. 8, 284–295 (2007).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Roth, S. Y., Denu, J. M. & Allis, C. D. Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 (2001).
Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R. & Young, R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007).
Saunders, A., Core, L. J. & Lis, J. T. Breaking barriers to transcription elongation. Nature Rev. Mol. Cell Biol. 7, 557–567 (2006).
Price, D. H. Poised polymerases: on your mark...get set...go! Mol. Cell 30, 7–10 (2008).
Eberhardy, S. R. & Farnham, P. J. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276, 48562–48571 (2001).
Eberhardy, S. R. & Farnham, P. J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 277, 40156–40162 (2002).
Bouchard, C., Marquardt, J., Bras, A., Medema, R. H. & Eilers, M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 23, 2830–2840 (2004).
Cowling, V. H., Chandriani, S., Whitfield, M. L. & Cole, M. D. A conserved Myc protein domain, MBIV, regulates DNA binding, apoptosis, transformation, and G2 arrest. Mol. Cell Biol. 26, 4226–4239 (2006).
Maruyama, K., Schiavi, S. C., Huse, W., Johnson, G. L. & Ruley, H. E. myc and E1A oncogenes alter the responses of PC12 cells to nerve growth factor and block differentiation. Oncogene 1, 361–367 (1987).
Spandidos, D. A. The effect of exogenous human ras and myc oncogenes in morphological differentiation of the rat pheochromocytoma PC12 cells. Int. J. Dev. Neurosci. 7, 1–4 (1989).
Hopewell, R. & Ziff, E. B. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner Max. Mol. Cell Biol. 15, 3470–3478 (1995).
Ribon, V., Leff, T. & Saltiel, A. R. c-Myc does not require max for transcriptional activity in PC-12 cells. Mol. Cell Neurosci. 5, 277–282 (1994).
Bentley, D. L. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17, 251–256 (2005).
Shatkin, A. J. Capping of eucaryotic mRNAs. Cell 9, 645–653 (1976).
Shuman, S. What messenger RNA capping tells us about eukaryotic evolution. Nature Rev. Mol. Cell Biol. 3, 619–625 (2002).
Schwer, B., Mao, X. & Shuman, S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 26, 2050–2057 (1998).
Moteki, S. & Price, D. Functional coupling of capping and transcription of mRNA. Mol. Cell 10, 599–609 (2002).
Komarnitsky, P., Cho, E. J. & Buratowski, S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 (2000).
Schroeder, S. C., Schwer, B., Shuman, S. & Bentley, D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14, 2435–2440 (2000).
Schroeder, S. C., Zorio, D. A., Schwer, B., Shuman, S. & Bentley, D. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13, 377–387 (2004).
Li, Z. et al. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl Acad. Sci. USA 100, 8164–8169 (2003).
Machida, Y. J., Hamlin, J. L. & Dutta, A. Right place, right time, and only once: replication initiation in metazoans. Cell 123, 13–24 (2005).
Felsher, D. W. & Bishop, J. M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc. Natl Acad. Sci. USA 96, 3940–3944 (1999).
Mai, S. et al. Chromosomal and extrachromosomal instability of the cyclin D2 gene is induced by Myc overexpression. Neoplasia 1, 241–252 (1999).
Cole, M. D. The myc oncogene: its role in transformation and differentiation. Ann. Rev. Gen. 20, 361–385 (1986).
Acknowledgements
This work was funded by National Institutes of Health research grants to M.D.C. and a Medical Research Council Career Development Award to V.H.C.
Author information
Authors and Affiliations
Corresponding author
Related links
Rights and permissions
About this article
Cite this article
Cole, M., Cowling, V. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat Rev Mol Cell Biol 9, 810–815 (2008). https://doi.org/10.1038/nrm2467
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2467
This article is cited by
-
Altered vitamin B12 metabolism in the central nervous system is associated with the modification of ribosomal gene expression: new insights from comparative RNA dataset analysis
Functional & Integrative Genomics (2023)
-
Identification of Hub Genes in Atypical Teratoid/Rhabdoid Tumor by Bioinformatics Analyses
Journal of Molecular Neuroscience (2020)
-
Emerging roles of Myc in stem cell biology and novel tumor therapies
Journal of Experimental & Clinical Cancer Research (2018)
-
Aluminum Trichloride Inhibited Osteoblastic Proliferation and Downregulated the Wnt/β-Catenin Pathway
Biological Trace Element Research (2017)
-
Missing link between microRNA and prostate cancer
Tumor Biology (2016)