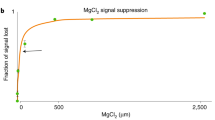
Abstract
Technologies for genome-wide analyses typically undergo a transition from a discovery phase to a scoring phase. In the discovery phase, the genomic universe is explored and all pertinent data are noted. In the scoring phase, relevant entities are screened to reveal groups of genes that are associated with specific biological processes or conditions. In this article, we propose that the transition from a discovery to a scoring phase is also essential, feasible and imminent for proteomics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Patterson, S. D. & Aebersold, R. H. Proteomics: the first decade and beyond. Nature Genet. 33, 311–323 (2003).
Steen, H. & Mann, M. The abc's (and xyz's) of peptide sequencing. Nature Rev. Mol. Cell Biol. 5, 699–711 (2004).
Andersen, J. S. et al. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 (2002).
Blagoev, B. et al. A proteomics strategy to elucidate functional protein–protein interactions applied to EGF signaling. Nature Biotechnol. 21, 315–318 (2003).
Bouwmeester, T. et al. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nature Cell Biol. 6, 97–105 (2004).
Brand, M. et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nature Struct. Mol. Biol. 11, 73–80 (2004).
Rosenblatt, K. P. et al. Serum proteomics in cancer diagnosis and management. Annu. Rev. Med. 55, 97–112 (2004).
Liotta, L. A., Ferrari, M. & Petricoin, E. Clinical proteomics: written in blood. Nature 425, 905 (2003).
Shiio, Y. et al. Quantitative proteomic analysis of Myc oncoprotein function. EMBO J. 21, 5088–5096 (2002).
Adams, M. D. et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252, 1651–1656 (1991).
Sachidanandam, R. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933 (2001).
LaBaer, J. & Ramachandran, N. Protein microarrays as tools for functional proteomics. Curr. Opin. Chem. Biol. 9, 14–19 (2005).
Desiere, F. et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 6, R9 (2005).
Keller, A., Nesvizhskii, A. I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Craig, R., Cortens, J. P. & Beavis, R. C. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 (2004).
Gygi, S. P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnol. 17, 994–999 (1999).
Schirle, M., Heurtier, M. A. & Kuster, B. Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography–tandem mass spectrometry. Mol. Cell. Proteomics 2, 1297–1305 (2003).
Washburn, M. P., Wolters, D. & Yates, J. R. III . Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnol. 19, 242–247 (2001).
Gavin, A. C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Kawashima, S., Ogata, H. & Kanehisa, M. AAindex: amino acid index database. Nucleic Acids Res. 27, 368–369 (1999).
Pan, S. et al. High-throughput proteome-screening for biomarker detection. Mol. Cell. Proteomics 4, 182–190 (2005).
Gerber, S. A., Rush, J., Stemman, O., Kirschner, M. W. & Gygi, S. P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl Acad. Sci. USA 100, 6940–6945 (2003).
Lu, Y., Bottari, P., Turecek, F., Aebersold, R. & Gelb, M. H. Absolute quantification of specific proteins in complex mixtures using visible isotope-coded affinity tags. Anal. Chem. 76, 4104–4111 (2004).
Lipton, M. S. et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl Acad. Sci. USA 99, 11049–11054 (2002).
Li, X. J. et al. A tool to visualize and evaluate data obtained by liquid chromatography–electrospray ionization-mass spectrometry. Anal. Chem. 76, 3856–3860 (2004).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Kuster, B., Schirle, M., Mallick, P. et al. Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol 6, 577–583 (2005). https://doi.org/10.1038/nrm1683
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm1683
This article is cited by
-
Existing and novel biomarkers for precision medicine in systemic sclerosis
Nature Reviews Rheumatology (2018)
-
Non-specific Digestion Artifacts of Bovine Trypsin Exemplified with Surrogate Peptides for Endogenous Protein Quantitation
Chromatographia (2018)