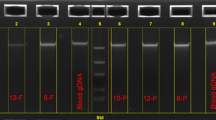
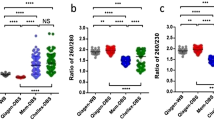
Abstract
Newborn screening (NBS) is a public-health genetic screening programme aimed at early detection and treatment of pre-symptomatic children affected by specific disorders. It currently involves protein-based assays and PCR to confirm abnormal results. We propose that DNA microarray technology might be an improvement over protein assays in the first stage of NBS. This approach has important advantages, such as multiplex analysis, but also has disadvantages, which include a high initial cost and the analysis/storage of large data sets. Determining the optimal technology for NBS will require that technical, public health and ethical considerations are made for the collection and extent of analysis of paediatric genomic data, for privacy and for parental consent.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Lipshutz, R. J., Fodor, S. P. A., Gingeras, T. R. & Lockart, D. J. High density synthetic oligonucleotide arrays. Nature Genet. 21, 20–24 (1999).
Wilgenbus, K. K. & Lichter, P. DNA chip technology ante portas. J. Mol. Med. 77, 761–768 (1999).
Ramaswamy, S. Translating cancer genomics into clinical oncology. N. Engl. J. Med. 350, 1814–1816, (2004).
Khoury, M. J., McCabe, L. L. & McCabe, E. R. Population screening in the age of genomic medicine. N. Engl. J. Med. 348, 50–58 (2003).
Wilson, J. M. G. & Junger, G. Principles and practice of screening for disease. Public health paper number 34 (World Health Organization, Geneva, 1968).
Rinaldo, P., Tortorelli, S. & Matern, D. Recent developments and new applications of tandem mass spectrometry in newborn screening. Curr. Opin. Pediatr. 16, 427–433 (2004).
Comeau, A. M. et al. Population-based newborn screening for genetic disorders when multiple mutation DNA testing is incorporated: a cystic fibrosis newborn screening model demonstrating increased sensitivity but more carrier detections. Pediatrics 113, 1573–1581 (2004).
Bhardwaj, U. et al. DNA diagnosis confirms hemoglobin deletion in newborn screen follow-up. J. Pediatr. 142, 346–348 (2003).
Pinto, G. et al. Follow-up of 68 children with congenital adrenal hyperplasia due to 21-hydroxylase deficiency: relevance of genotype for management. J. Clin. Endocrinol. Metab. 88, 2624–2633 (2003).
Green, N. S., Dolan, S. M. & Oinuma, M. Implementation of newborn screening for cystic fibrosis varies widely between states. Pediatrics 114, 515–516 (2004).
Hacia, J. G. & Collins, F. S. Mutational analysis using oligonucleotide microarrays. J. Med. Genet. 36, 730–766 (1999).
Cho, N. H. et al. Genotyping of 22 human papillomavirus types by DNA chip in Korean women: comparison with cytologic diagnosis. Am. J. Obstet. Gynecol. 188, 56–62 (2003).
Srivastava, M. et al. ANX7, a candidate tumor suppressor gene for prostate cancer. Proc. Natl Acad. Sci. USA 98, 4575–4580 (2001).
Primdahl, H. et al. Allelic imbalances in human bladder cancer: genome-wide detection with high-density single-nucleotide polymorphism arrays. J. Natl Cancer Inst. 94, 216–223 (2002).
Sellick, G. S., Garrett, C. & Houlston, R. S. A novel gene for neonatal diabetes maps to chromosome 10p12.1–p13. Diabetes 52, 2636–2638 (2003).
Mischler, E. H. et al. Cystic fibrosis newborn screening: impact on reproductive behavior and implications for genetic counseling. Pediatrics 102, 44–52 (1998).
Elsas, L. J. 2nd & Lai, K. The molecular biology of galactosemia. Genet. Med. 1, 40–48 (1998).
Grody, W. W. et al. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 3, 149–154 (2001).
European Society of Human Genetics. Data storage and DNA banking for biomedical research: technical, social and ethical issues. Eur. J. Hum. Genet. 1 (Suppl. 2), S8–S10 (2003).
Hoppe, C. et al. Gene interactions and stroke risk in children with sickle cell anemia. Blood 103, 2391–2396 (2004).
Clemens, C. J. & Davis, S. A. Minimizing false-positives in universal newborn hearing screening: a simple solution. Pediatrics 107, E29 (2001).
Fernandez-Burriel, M. & Rodriguez-Quinones, F. A simple method of screening for the common connexin-26 gene 35delG mutation in nonsyndromic neurosensory autosomal recessive deafness. Genet. Test. 7, 147–149 (2003)
Kenneson, A., Van Naarden Braun, K. & Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet. Med. 4, 258–274 (2002).
Fowler, K. B., Dahle, A. J., Boppana, S. B. & Pass, R. F. Newborn hearing screening: will children with hearing loss caused by congenital cytomegalovirus infection be missed? J. Pediatr. 135, 60–64 (1999).
Bailey, D. B. Newborn screening for fragile X syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 10, 3–10 (2004).
Kalman, L. et al. Mutations in genes required for T-cell development: IL7R, CD45, IL2RG, JAK3, RAG1, RAG2, ARTEMIS, and ADA and severe combined immunodeficiency: HuGE review. Genet Med. 6, 16–26 (2004).
Enns, G. M. et al. Molecular correlations in phenylketonuria: mutation patterns and corresponding biochemical and clinical phenotypes in a heterogeneous California population. Pediatr. Res. 46, 594–602 (1999).
Mendell, J. T. & Dietz, H. C. When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell 107, 411–414 (2001).
Inoue, K, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nature Genet. 36, 361–369 (2004).
Grody, W. W. Ethical issues raised by genetic testing with oligonucleotide microarrays. Mol. Biotechnol. 23, 127–138 (2003).
Clayton, E. W. Ethical, legal, and social implications of genomic medicine. N. Engl. J. Med. 349, 562–569 (2003).
Comeau, A. M. & Eaton, R. B. Successes of newborn screening programs. 295, 44–45 (2002).
European Society of Human Genetics. Population genetic screening programmes: technical, social and ethical issues. Eur. J. Hum. Genet. 11 (Suppl. 2), S5–S7 (2003).
Henn, W. Genetic screening with the DNA chip: a new Pandora's box? J. Med. Ethics 25, 200–203 (1999).
Wion, E. et al. Population-wide infant screening for HLA-based type 1 diabetes risk via dried blood spots from the public health infrastructure. Ann. NY Acad. Sci. 1005, 400–403 (2003).
Taub, J. W. et al. High frequency of leukemic clones in newborn screening blood samples of children with B-precursor acute lymphoblastic leukemia. Blood 99, 2992–2996 (2002).
Acknowledgements
We would like to thank Scott Grosse, Anne Marie Comeau and Harry Hannon for their valuable advice. The opinions stated here are entirely our own and do not necessarily reflect the positions of the March of Dimes or the NY State Department of Health. This work was in part supported by a National Human Genome Research Institute award to Thomas H. Murray.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
OMIM
FURTHER INFORMATION
AmpliChip CYP450 Test products page
National Newborn Screening and Genetic Resource Center
Statement of the American college of medical genetics on universal newborn hearing screening
Glossary
- ADRENAL HYPERPLASIA
-
An impairment of a key enzyme in the production of cortisol and aldosterone by the adrenal gland. It leads to impaired salt and fluid balance, with possible dehydration and shock, as well as the masculinization of sex organs in female infants.
- CONGENITAL HYPOTHYROIDISM
-
A condition in which insufficient thyroid hormone is produced. It can result from defects in the thyroid gland itself, or in other parts of the thyroid control system, including pituitary and hypothalamus.
- DUARTE MUTATION
-
One of two specific mutations in the galactose-1-phosphate uridyl transferase (GALT) gene, which is involved in the metabolism of galactose. When homozygous or when paired with another mutation in the GALT gene, this mutation is usually associated with mild clinical symptoms. Other GALT mutations have severe clinical effects.
- ISOELECTRIC FOCUSING
-
One of several electrophoretic procedures commonly used in newborn screening to identify different haemoglobins and their variants.
- REFLEX MUTATION ANALYSIS
-
A second DNA test performed to clarify the impact of mutations found when results from the first DNA analysis reveal mutations known to modify the effects of other gene mutations.
- TANDEM MASS SPECTROMETRY
-
An analytical system in which two linked mass spectrometers are used to measure small amounts of metabolites. The analytes are separated according to their mass and charge. By programing the instrument to only respond to certain masses, a high degree of specificity and sensitivity can be achieved.
- VAS DEFERENS
-
The proximal portion of the male reproductive tract through which sperm travel from the epididymis to the urethra.
Rights and permissions
About this article
Cite this article
Green, N., Pass, K. Neonatal screening by DNA microarray: spots and chips. Nat Rev Genet 6, 147–151 (2005). https://doi.org/10.1038/nrg1526
Issue Date:
DOI: https://doi.org/10.1038/nrg1526
This article is cited by
-
Whole genome microarray analysis, from neonatal blood cards
BMC Genetics (2009)
-
A Systematic Review of the Effects of Disclosing Carrier Results Generated Through Newborn Screening
Journal of Genetic Counseling (2008)
-
Reduction of the false‐positive rate in newborn screening by implementation of MS/MS‐based second‐tier tests: The Mayo Clinic experience (2004–2007)
Journal of Inherited Metabolic Disease (2007)
-
Estimation of the total number of disease‐causing mutations in ornithine transcarbamylase (OTC) deficiency. Value of the OTC structure in predicting a mutation pathogenic potential
Journal of Inherited Metabolic Disease (2007)
-
Storage policies and use of the Danish Newborn Screening Biobank
Journal of Inherited Metabolic Disease (2007)