Key Points
-
Programmed cell death, or apoptosis, is ubiquitous in animals, where it has an important function in development and homeostasis.
-
Drosophila melanogaster is an attractive system in which to study apoptosis because it occurs throughout the fly life cycle and in response to a number of insults that are relevant to human disease. In addition, where studied, cell death in flies and mammals involves similar machinery and mechanisms of regulation.
-
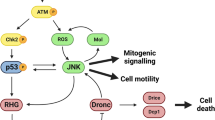
Caspases are the core of the cell-death machinery, and much of the regulation of cell death revolves around controlling their activation and their activity once they are activated. Inhibition of DIAP1, an anti-apoptotic IAP family member essential for cell survival, is an important site of pro-apoptotic protein action in D. melanogaster.
-
Several outstanding questions remain in the study of cell death in D. melanogaster. For example, how do the known components of the canonical cell-death pathway interact and regulate each other's activities? How many core components remain to be discovered? And which other non-canonical cell-death pathways are involved? These, and other issues can be studied using the wide range of sophisticated genetic and molecular techniques that have become available for this organism.
-
F2 loss-of-function screens have led to the identification of important cell-death regulators. New tools have increased the power of these screens to rapidly identify interesting genes.
-
Clone-based screens that involve tissue-specific loss-of-function provide an approach to identifying cell-death regulators in any tissue and at any life stage.
-
The dominant modifier screen provides a method for rapidly identifying components of a specific death-signalling pathway. These are identified as enhancers or suppressors of a dominant phenotype that has been created to produce a sensitized genetic background.
-
Cell-culture-based RNAi screens can identify genes that are required for cell survival or cell death in different contexts.
Abstract
Cell death is ubiquitous in metazoans and involves the action of an evolutionarily conserved process known as programmed cell death or apoptosis. In Drosophila melanogaster, it is now uniquely possible to screen for genes that determine the fate — life or death — of any cell or population of cells during development and in the adult. This review describes these genetic approaches and the key insights into cell-death mechanisms that have been obtained, as well as the outstanding questions that these techniques can help to answer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kerr, J. F. R., Wyllie, A. H. & Curie, A. R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 (1972).
Raff, M. C. Social controls on cell survival and cell death. Nature 356, 397–400 (1992).
Vaux, D. L. & Korsmeyer, S. J. Cell death in development. Cell 96, 245–254 (1999).
Baehrecke, E. H. How death shapes life during development. Nature Rev. Mol. Cell Biol. 3, 779–787 (2002).
Green, D. R. & Evan, G. I. A matter of life and death. Cancer Cell 1, 19–30 (2002).
Benedict, C. A., Norris, P. S. & Ware, C. F. To kill or be killed: viral evasion of apoptosis. Nature Immunol. 3, 1013–1018 (2002).
James, E. R. & Green, D. R. Manipulation of apoptosis in the host-parasite interaction. Trends Parasitol. 20, 280–287 (2004).
Opferman, J. T. & Korsmeyer, S. J. Apoptosis in the development and maintenance of the immune system. Nature Immunol. 4, 410–415 (2003).
Hochachka, P. W. & Lutz, P. L. Mechanism, origin and evolution of anoxia tolerance in animals. Comp. Biochem. Physiol. B 130, 435–459 (2001).
Vaux, D. L. & Flavell, R. A. Apoptosis genes and autoimmunity. Curr. Opin. Immunol. 12, 719–724 (2000).
Badley, A. D., Roumier, T., Lum, J. J. & Kroemer, G. Mitochondrion-mediated apoptosis in HIV-1 infection. Trends Pharmacol. Sci. 24, 298–305 (2003).
Muqit, M. M. & Feany, M. B. Modelling neurodegenerative diseases in Drosophila: a fruitful approach? Nature Rev. Neurosci. 3, 237–243 (2002).
Bonini, N. M. & Fortini, M. E. Human neurodegenerative disease modeling using Drosophila. Annu. Rev. Neurosci. 26, 627–656 (2003).
Richardson, H. & Kumar, S. Death to flies: Drosophila as a model system to study programmed cell death. J. Immunol. Methods 265, 21–38 (2002).
Bernards, A. & Hariharan, I. K. Of flies and men — studying human disease in Drosophila. Curr. Opin. Genet. Dev. 11, 274–278 (2001).
Vernooy, S. Y. et al. Cell death regulation in Drosophila: conservation of mechanism and unique insights. J. Cell Biol. 150, F69–76 (2000).
Aravind, L., Dixit, V. M. & Koonin, E. V. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science 291, 1279–1284 (2001).
Robinson, A. S., Franz, G. & Atkinson, P. W. Insect transgenesis and its potential role in agriculture and human health. Insect Biochem. Mol. Biol. 34, 113–120 (2004).
St Johnston, D. The art and design of genetic screens: Drosophila melanogaster. Nature Rev. Genet. 3, 176–188 (2002).
Adams, M. D. & Sekelsky, J. J. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nature Rev. Genet. 3, 189–198 (2002).
McGuire, S. E., Roman, G. & Davis, R. L. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 20, 384–391 (2004).
Boutros, M. et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832–835 (2004). Describes the results of a genome-scale, cell-culture-based RNAi screen for genes required for cell survival, growth or metabolism.
Xu, P., Vernooy, S. Y., Guo, M. & Hay, B. A. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13, 790–795 (2003).
Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 (2003). References 23 and 24 report the identification of miRNA cell-death inhibitors from dominant modifier and overexpression screens, respectively.
Wu, S., Huang, J., Dong, J. & Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 (2003).
Pantalacci, S., Tapon, N. & Leopold, P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nature Cell Biol. 5, 921–927 (2003).
Harvey, K. F., Pfleger, C. M. & Hariharan, I. K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 (2003).
Jia, J., Zhang, W., Wang, B., Trinko, R. & Jiang, J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514–2519 (2003).
Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C. & Halder, G. Hippo, the Drosophila homologue of the MST1/2 kinases, promotes cell proliferation arrest and apoptosis in the Salvador/Warts pathway. Nature Cell Biol. 5, 914–920 (2003). References 25–29 report the identification of HIP, an Mst family kinase, using clone-based screens for negative regulators of cell growth. HIP restricts cell growth and promotes apoptosis.
Hipfner, D. R. & Cohen, S. M. The Drosophila Sterile-20 kinase Slik controls cell proliferation and apoptosis during imaginal disc development. PLOS Biol. 1, E35 (2003).
Arama, E., Agapite, J. & Steller, H. Caspase activity and a specific cytochrome c are required for sperm differentiation in Drosophila. Dev. Cell 4, 687–697 (2003).
Huh, J. R. et al. Multiple apoptotic caspase caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLOS Biol. 2, 43–53 (2004).
Huh, J. R., Guo, M. & Hay, B. A. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical caspase Dronc in a nonapoptotic role. Curr. Biol. 14, 1262–1266 (2004).
Geisbrecht, E. R. & Montell, D. J. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell 118, 111–125 (2004). References 31–34 describe non-apoptotic roles for cell-death activators in D. melanogaster.
Degterev, A., Boyce, M. & Yuan, J. A decade of caspases. Oncogene 22, 8543–8567 (2003).
Adams, J. M. Ways of dying: mutiple pathways to apoptosis. Genes Dev. 17, 2481–2495 (2003).
Jiang, X. & Wang, X. Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 (2004).
Weil, M. et al. Constitutive expression of the machinery for programmed cell death. J. Cell Biol. 133, 1053–1059 (1996).
Wang, S. L., Hawkins, C. J., Yoo, S. J., Muller, H. A. & Hay, B. A. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98, 453–463 (1999).
Metzstein, M. M., Stanfield, G. M. & Horvitz, H. R. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 14, 410–416 (1998).
Yan, N. et al. Structural, biochemical, and functional analysis of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol. Cell 15, 999–1006 (2004).
Fischer, U., Janicke, R. U. & Schulze-Osthoff, K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10, 76–100 (2003).
Shi, Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9, 459–470 (2002).
Salvesen, G. S. & Duckett, C. S. IAP proteins: blocking the road to death's door. Nature Rev. Mol. Cell Biol. 3, 401–410 (2002).
Igaki, T., Yamamoto-Goto, Y., Tokushige, N., Kanda, H. & Miura, M. Down-regulation of DIAP1 triggers a novel Drosophila cell death pathway mediated by Dark and DRONC. J. Biol. Chem. 277, 23103–23106 (2002).
Muro, I., Hay, B. A. & Clem, R. J. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J. Biol. Chem. 277, 49644–49650 (2002).
Rodriguez, A., Chen, P., Oliver, H. & Abrams, J. M. Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J. 21, 2189–2197 (2002).
Zimmermann, K. C., Ricci, J. E., Droin, N. M. & Green, D. R. The role of ARK in stress-induced apoptosis in Drosophila cells. J. Cell Biol. 156, 1077–1087 (2002).
Muro, I., Monser, K. & Clem, R. J. Mechanism of Dronc activation in Drosophila cells. J. Cell Sci. 117, 5035–5041 (2004).
Hay, B. A., Wassarman, D. A. & Rubin, G. M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83, 1253–1262 (1995). This classic paper describes the first dominant modifier screen for cell-death inhibitors, which identified DIAP1 in flies.
Hay, B. A. Understanding IAP function and regulation: a view from Drosophila. Cell Death Differ. 7, 1045–1056 (2000).
Bergmann, A., Yang, A. Y. & Srivastava, M. Regulators of IAP function: coming to grip with the grim reaper. Curr. Opin. Cell Biol. 15, 717–724 (2003).
Kuranaga, E. et al. Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nature Cell Biol. 4, 705–710 (2002).
Kanda, H. & Miura, M. Regulatory roles of JNK in programmed cell death. J. Biochem. (Tokyo) 136, 1–6 (2004).
Yoo, S. J. et al. Apoptosis inducers Hid, Rpr and Grim negatively regulate levels of the caspase inhibitor DIAP1 by distinct mechanisms. Nature Cell Biol. 4, 416–424 (2002).
Holley, C. L., Olson, M. R., Colon-Ramos, D. A. & Kornbluth, S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nature Cell Biol. 4, 439–444 (2002).
Claveria, C., Caminero, E., Martinez, A. C., Campuzano, S. & Torres, M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 21, 3327–3336 (2002).
Chen, P., Ho, S.-I., Shi, Z. & Abrams, J. M. Bifunctional killing activity encoded by conserved reaper proteins. Cell Death Differ. 11, 704–713 (2004).
Huh, J. R. & Hay, B. A. Sculpture of a fly's head. Nature 418, 926–928 (2002).
Kumar, S. & Cakouros, D. Transcriptional control of the core cell-death machinery. Trends Biochem. 29, 193–199 (2004).
Lohmann, I., McGinnis, N., Bodmer, M. & McGinnis, W. The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell 110, 457–466 (2002). This paper reports the identification of RPR as a target of the transcription factor DFD in a screen for mutations that display aspects of the Deformed embryonic phenotype.
Brodsky, M. H. et al. Drosophila p53 binds a damage response element at the reaper locus. Cell 101, 103–113 (2000).
Cakouros, D., Daish, T. J. & Kumar, S. Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc, regulating its expression in specific tissues. J. Cell Biol. 165, 631–640 (2004).
Franc, N. C. Phagocytosis of apoptotic cells in mammals, Caenorhabditis elegans and Drosophila melanogaster: molecular mechanisms and physiological consequences. Front Biosci. 7, d1298–1313 (2002).
Mergliano, J. & Minden, J. S. Caspase-independent cell engulfment mirrors cell death pattern in Drosophila embryos. Development 130, 5779–5789 (2003). This paper identifies a RPR-, HID-, GRIM- and p35-independent step in cell death during fly embryogenesis.
Igaki, T. & Miura, M. Role of bcl-2 family members in invertebrates. Biochim. Biophys. Acta 1644, 73–81 (2004).
McCall, K. Eggs over easy: cell death in the Drosophila ovary. Dev. Biol. 274, 3–14 (2004).
Baehrecke, E. H. Autophagic programmed cell death in Drosophila. Cell Death Differ. 10, 940–945 (2003).
Driscoll, M. & Gerstbrein, B. Dying for a cause: invertebrate genetics takes on human neurodegeneration. Nature Rev. Genet. 4, 181–194 (2003).
Jackson, G. R. et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21, 633–642 (1998).
Hsu, C. D. et al. Limited role of developmental programmed cell death pathways in Drosophila norpA retinal degeneration. J. Neurosci. 24, 500–507 (2004).
McLaughlin, B. The kinder side of killer proteases: caspase activation contributes to neuroprotection and CNS remodeling. Apoptosis 9, 111–121 (2004).
Schwerk, C. & Schulze-Osthoff, K. Non-apoptotic functions of caspases in cellular proliferation and differentiation. Biochem. Pharmacol. 66, 1453–1458 (2003).
Wolff, T. & Ready, D. F. Cell death in normal and rough eye mutants of Drosophila. Development 113, 825–839 (1991). Reports the first cell death mutants to be identified in D. melanogaster.
White, K. et al. Genetic control of programmed cell death in Drosophila. Science 264, 677–683 (1994). This classic paper described the deficiency screen that led to the identification of rpr , and ultimately hid and grim.
Grether, M. E., Abrams, J. M., Agapite, J., White, K. & Steller, H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9, 1694–1708 (1995).
Chen, P., Nordstrom, W., Gish, B. & Abrams, J. M. grim, a novel cell death gene in Drosophila. Genes Dev. 10, 1773–1782 (1996).
Peterson, C., Carney, G. E., Taylor, B. J. & White, K. reaper is required for neuroblast apoptosis during Drosophila development. Development 128, 1467–1476 (2002).
Hiesinger, P. R. & Bellen, H. J. Flying in the face of total disruption. Nature Genet. 36, 211–212 (2004).
Jassim, O. W., Fink, J. L. & Cagan, R. L. Dmp53 protects the Drosophila retina during a developmentally regulated DNA damage response. EMBO J. 22, 5622–5632 (2003).
Foe, V. E. & Alberts, B. M. Reversible chromosome condensation induced in Drosophila embryos by anoxia: visualization of interphase nuclear organization. J. Cell Biol. 100, 1623–1636 (1985).
Teodoro, R. O. & O'Farrell, P. H. Nitric oxide-induced suspended animation promotes survival during hypoxia. EMBO J. 22, 580–587 (2003).
Tapon, N. et al. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 (2002).
Kango-Singh, M. et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719–5730 (2002).
Justice, R. W., Zilian, O., Woods, D. F., Noll, M. & Bryant, P. J. The Drosophila tumor-suppressor gene warts encodes a homolog of human myotonic-dystrophy kinase and is required for the control of cell-shape and proliferation. Genes Dev. 9, 534–546 (1995).
Xu, T. A., Wang, W. Y., Zhang, S., Stewart, R. A. & Yu, W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 (1995).
Hay, B. A. & Guo, M. Coupling cell growth, proliferation, and death: hippo weighs in. Dev. Cell 5, 361–363 (2003).
Kimura, K. -I., Kodama, A., Hayasaka, Y. & Ohta, T. Activation of the cAMP/PKA signaling pathway is required for post-ecdysial cell death in wing epidermal cells of Drosophila. Development 131, 1597–1606 (2004). This paper uses differential GFP labelling of live versus dead cells to visualize the kinetics and genetic requirements for cell death in the adult fly wing.
Perrimon, N., Engstrom, L. & Mahowald, A. P. Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics 121, 333–352 (1989).
Martin, S. G., Leclerc, V., Smith-Litiere, K. & St. Johnston, D. The identification of novel genes required for Drosophila anteroposterior axis formation in a germline clone screen using GFP-Staufen. Development 130, 4201–4215 (2003).
Luschnig, S. et al. An F1 genetic screen for maternal-effect mutations affecting embryonic pattern formation in Drosophila melanogaster. Genetics 167, 325–342 (2004).
Bellotto, M. et al. Maternal-effect loci involved in Drosophila oogenesis and embryogenesis: P element-induced mutations on the third chromosome. Int. J. Dev. Biol. 46, 149–157 (2002).
Hays, R., Wickline, L. & Cagan, R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nature Cell Biol. 4, 425–431 (2002).
Hay, B. A., Maile, R. & Rubin, G. M. P element insertion-dependent gene activation in the Drosophila eye. Proc. Natl Acad. Sci. USA 94, 5195–5200 (1997).
Rorth, P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl Acad. Sci. USA 93, 12418–12422 (1996).
Vernooy, S. Y. et al. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr. Biol. 12, 1164–1168 (2002).
Wing, J. P. et al. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nature Cell Biol. 4, 451–456 (2002).
Ryoo, H. D., Bergmann, A., Gonen, H., Ciechanover, A. & Steller, H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nature Cell Biology 4, 432–438 (2002).
Hipfner, D. R., Weigmann, K. & Cohen, S. M. The bantam gene regulates Drosophila growth. Genetics 161, 1527–1537 (2002).
Rong, Y. S. Gene targeting by homologous recombination: a powerful addition to the genetic arsenal for Drosophila geneticists. Biochem. Biophys. Res. Comm. 297, 1–5 (2002).
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349 (2004).
Foley, E. & O'Farrell, P. H. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLOS Biol. 2, e203 (2004). A cell-culture-based reporter system was used to identify regulators of the innate immune response, which requires the caspase DREDD. A potential negative regulator of DREDD activity, DNR1, was identified.
Wu, R. Z., Bailey, S. N. & Sabatini, D. M. Cell-biological applications of transfected-cell microarrays. Trends Cell Biol. 12, 485–488 (2002).
Eschalier, G. Drosophila Cells in Culture (Academic Press, New York, 1997).
Chen, P., Rodriguez, A., Erskine, R., Thach, T. & Abrams, J. M. Dredd, a novel effector of the apoptosis activators Reaper, Grim, and Hid in Drosophila. Dev. Biol. 201, 202–216 (1998).
Hultmark, D. Drosophila immunity: paths and patterns. Curr. Opin.Immunol. 15, 12–19 (2003).
Lockshin, R. A. & Zakeri, Z. Caspase-independent cell death? Oncogene 23, 2766–2773 (2004).
Jaattela, M. Multiple cell death pathways as regulators of tumour initiation and progression. Oncogene 23, 2746–2756 (2004).
Reiter, L. T., Potocki, L., Chien, S., Gribskov, M. & Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11, 1114–1125 (2001).
Giot, L. et al. A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 (2003). This paper is remarkable for its failure to identify any of the known physical interactions between components of the core apoptotic machinery, thereby highlighting the limitations of this approach.
Vucic, D., Kaiser, W. J. & Miller, L. K. Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol. Cell Biol. 18, 3300–3309 (1998).
Acknowledgements
Work in the authors' laboratories is supprted by NIH (National Institutes of Health) grants. We apologize to authors whose work could not be cited directly owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez
OMIM
Swiss-Prot
FURTHER INFORMATION
Glossary
- RNAi-MEDIATED KNOCKDOWN
-
A phenomenon in which the expression of a gene is temporarily inhibited ('knocked down') when a complementary dsRNA molecule is introduced into the organism.
- SATURATION
-
A screen that is designed to induce at least one mutation in every gene is said to have been carried out to saturation.
- BACULOVIRUS CASPASE INHIBITOR P35
-
Baculoviruses are large DNA viruses that infect arthropods. p35 is a baculovirus-encoded protein that inhibits cell death by acting as a suicide substrate for many caspases.
- RNA INTERFERENCE (RNAi)
-
A form of post-transcriptional gene silencing, in which dsRNA induces degradation of the homologous endogenous transcripts, mimicking the effect of the reduction, or loss, of gene activity.
- EPISTASIS
-
When the phenotype associated with mutation of a gene (A) is masked by mutation in a second gene (B), B is said to be epistatic to A. In a switch pathway (a pathway in which the output is one of two states, often developmental fates), such an observation would indicate that genes A and B act in the same pathway, and that A acts through B.
- SENSITIZED GENETIC BACKGROUND
-
A genetic background in which modest (twofold) changes in the dose of pathway components produce a phenotype that would not be observed in a wild-type background.
- DOMINANT MODIFIER SCREEN
-
A signalling pathway is hyperactivated or partially deactivated in a specific tissue. These flies are often sensitive to modest changes in the levels of pathway components (heterozygosity) that would otherwise not result in a visible phenotype — but only in the specific tissue that is targeted.
- NURSE CELLS
-
Female germline-derived cells that support the development of the oocyte. Nurse cells are interconnected to each other and to the developing oocyte through intercellular bridges that facilitate transport of RNA and protein into the growing oocyte.
- IMAGINAL DISC
-
An epithelial sheet of cells that occurs as a sac-like infolding of the epithelium in the larva. Small groups of imaginal disc founder cells arise in the embryo. They continue to divide until pupation, when they differentiate into many adult structures (wings, legs, eyes, antennae and genitalia).
- SYNCYTIUM
-
A multinucleate cell in which the nuclei are not separated by cell membranes.
- AUTOPHAGY
-
In autophagic cell death, as opposed to apoptotic cell death, the cell is degraded largely from within, with little or no help from phagocytes. Bulk cytoplasm and organelles are sequestered within double-membrane-bound vesicles. These ultimately fuse with the lysosome and their contents are degraded.
- SPERMATID
-
A post-meiotic haploid male germ cell.
- BORDER CELLS
-
A small group of specialized somatic follicle cells. They delaminate from the follicular epithelium, invade the underlying germline tissue and migrate towards the oocyte.
- MITOTIC RECOMBINATION
-
A crossover between two homologous dsDNA molecules that leads to a physical exchange of DNA and genetic information. This recombination occurs frequently during meiosis, but is relatively rare during mitosis. As a consequence of mitotic recombination, cells can undergo a 'loss of heterozygosity' or gene conversion.
- BALANCED STOCK
-
A stock that carries a lethal mutation on one chromosome homologue, and a balancer chromosome on the other. A balancer chromosome carries multiple inversions that prevent recombination with the lethal-bearing chromosome, a recessive lethal mutation and a dominant marker. Matings between balanced lethal flies produce only balanced lethal adult progeny — a stable stock.
- ISOGENIC
-
Cells or organisms that are derived from the same parent and therefore have almost identical genomes.
- GERMLINE CLONE SCREEN
-
A genetic screen in which clones of homozygous-mutant germline tissue are produced in adult females. Oocytes and eggs derived from these clones (which can be distinguished from those derived from heterozygous germline tissue in several ways) can be examined for phenotypes during oogenesis and embryogenesis.
- P-ELEMENT
-
A member of a family of transposable elements that are widely used as the basis of tools for mutating and manipulating the genome of Drosophila melanogaster.
- CELL MICROARRAYS
-
Cells are plated directly onto a slide containing thousands of microarrayed spots of DNA. Cells landing on these spots are transfected with the arrayed plasmids, and can then be scored in various assays.
Rights and permissions
About this article
Cite this article
Hay, B., Huh, J. & Guo, M. The genetics of cell death: approaches, insights and opportunities in Drosophila. Nat Rev Genet 5, 911–922 (2004). https://doi.org/10.1038/nrg1491
Issue Date:
DOI: https://doi.org/10.1038/nrg1491
This article is cited by
-
Phylogenetic analysis of the caspase family in bivalves: implications for programmed cell death, immune response and development
BMC Genomics (2021)
-
Toll signaling promotes JNK-dependent apoptosis in Drosophila
Cell Division (2020)
-
Exploitation of Drosophila Choriogenesis Process as a Model Cellular System for Assessment of Compound Toxicity: the Phloroglucinol Paradigm
Scientific Reports (2020)
-
Apoptosis inhibition mitigates aging effects in Drosophila melanogaster
Genetica (2020)
-
Snail modulates JNK-mediated cell death in Drosophila
Cell Death & Disease (2019)