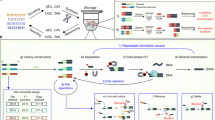
Abstract
Large-scale genome, transcriptome and proteome projects are rapidly producing new information, databases and physical resources that need to be delivered to the biomedical community in a quick and efficient way. Information and databases can be made available online but there was, until recently, no standardized means of widely distributing physical resources. However, the technology required is now available and the way laboratory researchers work might never be the same again.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Lucky, R. The quickening of science communication. Science 289, 259 (2000).
The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature 409, 685–690 (2001)
The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I & II Team. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573 (2002).
Kawai, J. & Hayashizaki, Y. DNA book. Genome Res. 13, 1488–1495 (2003).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Waterston, R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Burgoyne, L. Solid medium and method for DNA storage. US Patent 5,496,562 (1996).
Hayashizaki, Y. & Kawai, J. (eds.). RIKEN Mouse Genome Encyclopedia DNABook (RIKEN Genomic Sciences Center, Yokohama, 2003).
Hayashizaki, Y., Kawai, J. & Kanehisa, M. (eds.). RIKEN Human cDNA Encyclopedia Metabolome DNABook (RIKEN Genomic Sciences Center, Yokohama, 2003).
Kikuchi, S. et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379 (2003).
Seki, M. et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145 (2002).
Yamada, K. et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846 (2003).
Talbot, W. S. et al. Zebrafish mutations and functional analysis of the vertebrate genome. Genes Dev 14, 755–762 (2000).
Robert, K. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev. Dyn. 225, 384–391 (2002).
Satou, Y. et al. A cDNA resource from the basal chordate Ciona intestinalis. Genesis 33, 153–154 (2002).
Rubin, G. A Drosophila complementary DNA resource. Science 287, 2222–2224 (2000).
Okazaki, Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573 (2002).
Mammalian Gene Collection (MGC) Program Team. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA 99, 16899–16903 (2002).
The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Sasaki, T. The genome sequence and structure of rice chromosome 1. Nature 420, 312–316 (2002).
Gardner, M. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002).
Dehal, P. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157–2167 (2002).
Robert, H. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129–149 (2002).
Twigger, S. Rat Genome Database (RGD): mapping disease onto the genome. Nucl. Acids Res 30, 125–128 (2002).
IKirkness, E. F. The dog genome: survey sequencing and comparative analysis. Science 301, 1898–1903 (2003).
Acknowledgements
We thank S. Fukuda, K. Imamura, K. Imotani, M. Kanamori, T. Kasukawa, S. Katayama, F. Kawabata, T. Kawashima, Y. Kitazume, M. Nakamura, H. Sano, T. Sasaki, Y. Sumita, H. Suzuki, Y. Takeda and Y. Chen Choo for their technical support; T. Hayashi, H. Himei, H. Isaka, Y. Shigemoto, A. Suzuki, K. Suzuki, A. Tomotsugu and H. Torigoe for their secretarial support; and K.K. Dnaform for their self-financed cooperation. We especially thank J. Mattick and J. Gough for their editorial assistance. We also thank RIKEN executives, and the members of Yokohama Research Promotion Division and Technology Transfer and Research Coordination Division for administrative support. This work was supported by research grants from the Ministry of Education and Culture (for the RIKEN Genome Exploration Research Project), the Sports, Science and Technology Department of the Japanese Government (to Y.H.), and from the Japan Science and Technology Corporation (to Y.H. for Preventure).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- DNA PRINTER
-
A machine that is used for spotting DNA on substrate such as paper.
- FTA CARD
-
A chemically treated filter paper that is designed for the collection and storage of biological samples at room temperature for subsequent DNA analysis.
- FULL-LENGTH CDNA CLONES
-
Clones containing the entire fragments of complementary DNAs of mRNA.
- LAB-ON-A-CHIP
-
The technology that is used to perform a combination of analysis on a single miniaturized devise for biological and clinical analysis.
- TRANSCRIPTOME
-
The full complement of activated genes, mRNAs or transcripts in a particular tissue at a particular time.
Rights and permissions
About this article
Cite this article
Hayashizaki, Y., Kawai, J. A new approach to the distribution and storage of genetic resources. Nat Rev Genet 5, 223–228 (2004). https://doi.org/10.1038/nrg1296
Issue Date:
DOI: https://doi.org/10.1038/nrg1296
This article is cited by
-
DNA banking for plant breeding, biotechnology and biodiversity evaluation
Journal of Plant Research (2007)