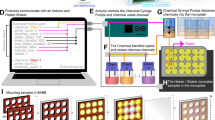
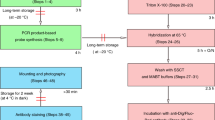
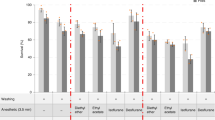
Abstract
Antibody staining is a vital technique for studying the development of many model organisms, including Drosophila. Reliable protocols have long been available for antibody staining of Drosophila whole-mount embryos and dissected larvae. By contrast, methods for staining whole larvae have rarely been reported, are unreliable, and fail to work on large third-instar larvae. This has become a major limitation to understanding the role of multitissue interactions such as neural circuit formation and cell metastasis. We have modified existing embryo protocols to develop a reliable method for antibody staining of whole Drosophila larvae of any developmental stage. The procedure consists of a bleach wash, enzymatic digestion, first fixation, 'cracking', second fixation, (optional) Proteinase K (Pro-K) or sonication treatment, antibody staining, clearing, and mounting. The method takes longer than typical antibody stains of dissected larval tissues—12 or 16 d, depending on the size of the larvae, compared with 2–3 d for embryos or dissected tissue stains—but time is saved by eliminating the need for larval dissections and by allowing hundreds of larvae to be batch-processed. The method also works well for staining embryos, even late-stage embryos with cuticles, allowing characterization from early embryogenesis to the end of larval development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
20 April 2017
In the version of this article initially published, two concentrations for Triton X-100 detergent were given in the blocking buffer ingredient list—0.3% is the correct concentration. The error has been corrected in the HTML and PDF versions of the article.
References
Nern, A., Pfeiffer, B.D. & Rubin, G.M. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967–E2976 (2015).
Chung, K. & Deisseroth, K. CLARITY for mapping the nervous system. Nat. Methods 10, 508–513 (2013).
Mitchison, T.J. & Sedat, J. Localization of antigenic determinants in whole Drosophila embryos. Dev. Biol. 99, 261–264 (1983).
Doe, C.Q. Neural stem cells: balancing self-renewal with differentiation. Development 135, 1575–1587 (2008).
Homem, C.C., Repic, M. & Knoblich, J.A. Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 16, 647–659 (2015).
Kohwi, M. & Doe, C.Q. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823–838 (2013).
Li, X., Chen, Z. & Desplan, C. Temporal patterning of neural progenitors in Drosophila. Curr. Top. Dev. Biol. 105, 69–96 (2013).
Parton, R.M., Valles, A.M., Dobbie, I.M. & Davis, I. Drosophila larval fillet preparation and imaging of neurons. Cold Spring Harb. Protoc. 2010 (2010).
Clark, M.Q., McCumsey, S.J., Lopez-Darwin, S., Heckscher, E.S. & Doe, C.Q. Functional genetic screen to identify interneurons governing behaviorally distinct aspects of Drosophila larval motor programs. G3 (Bethesda) 6, 2023–2031 (2016).
Patel, N.H. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 44, 445–487 (1994).
Arata, Y., Lee, J.Y., Goldstein, B. & Sawa, H. Extracellular control of PAR protein localization during asymmetric cell division in the C. elegans embryo. Development 137, 3337–3345 (2010).
Copf, T., Rabet, N., Celniker, S.E. & Averof, M. Posterior patterning genes and the identification of a unique body region in the brine shrimp Artemia franciscana. Development 130, 5915–5927 (2003).
Freeman, J.A., Cheshire, L.B. & MacRae, T.H. Epithelial morphogenesis in developing Artemia: the role of cell replication, cell shape change, and the cytoskeleton. Dev. Biol. 152, 279–292 (1992).
Fidler, A., Bouley, L. & Wawersik, M. Sonication-facilitated immunofluorescence staining of late-stage embryonic and larval Drosophila tissue in situ. J. Vis. Exp. 90, e51528 (2014).
Duerr, J.S. Immunohistochemistry. WormBook 19, 1–61 (2006).
Ott, S.R. Confocal microscopy in large insect brains: zinc-formaldehyde fixation improves synapsin immunostaining and preservation of morphology in whole-mounts. J. Neurosci. Methods 172, 220–230 (2008).
Horiuchi, D. et al. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr. Biol. 17, 1313–1317 (2007).
Pfeiffer, B.D. et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715–9720 (2008).
Acknowledgements
We thank E.S. Heckscher for her inspiration to begin this work, and B. Bowerman, K. Sugioka, and C.-H. Chuang for their help on the C. elegans protocols. This work was supported by the Howard Hughes Medical Institute, where C.Q.D. is an investigator.
Author information
Authors and Affiliations
Contributions
L.M. conceived and performed all experiments, collected all data, and wrote the first draft of the paper. C.Q.D. assembled the figures and provided comments on the text.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Manning, L., Doe, C. Immunofluorescent antibody staining of intact Drosophila larvae. Nat Protoc 12, 1–14 (2017). https://doi.org/10.1038/nprot.2016.162
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.162
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.