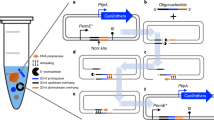
Abstract
PCR-based gene-targeting approaches have increased the speed of gene function analyses in ascomycetous fungi, for example, in the diploid human fungal pathogen Candida albicans. Here we describe a protocol that utilizes Rapid-PCR to amplify all cassettes available with the previously reported pFA modules. With this protocol, sufficient quantities of any cassette for use in C. albicans transformation experiments can be reliably generated in 25–50 min using either of the two alternative optimized amplification conditions; cassette amplification by standard PCR methods typically takes 3–4 h and is likely to require optimization of amplification conditions for each cassette. Transformants that appear 2–4 d after transformation can be rapidly identified using Rapid-PCR on whole cells, eliminating the need for genomic DNA extraction. In total, less than a week is required for the deletion of one allele in C. albicans. Repeating this procedure will result in the generation of homozygous mutant strains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hasty, P., Rivera-Perez, J. & Bradley, A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 11, 5586–5591 (1991).
Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F. & Cullin, C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329–3330 (1993).
Wach, A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12, 259–265 (1996).
Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793–1808 (1994).
Bahler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998).
Wilson, R.B., Davis, D. & Mitchell, A.P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868–1874 (1999).
Taroncher-Oldenburg, G. & Stephanopoulos, G. Targeted, PCR-based gene disruption in cyanobacteria: inactivation of the polyhydroxyalkanoic acid synthase genes in Synechocystis sp. PCC6803. Appl. Microbiol. Biotechnol. 54, 677–680 (2000).
Wendland, J., Ayad-Durieux, Y., Knechtle, P., Rebischung, C. & Philippsen, P. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242, 381–391 (2000).
Wendland, J. PCR-based methods facilitate targeted gene manipulations and cloning procedures. Curr. Genet. 44, 115–123 (2003).
Nielsen, M.L., Albertsen, L., Lettier, G., Nielsen, J.B. & Mortensen, U.H. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43, 54–64 (2006).
Winzeler, E.A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
Gietz, R.D. & Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Fairhead, C., Llorente, B., Denis, F., Soler, M. & Dujon, B. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using 'split-marker' recombination. Yeast 12, 1439–1457 (1996).
Ninomiya, Y., Suzuki, K., Ishii, C. & Inoue, H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101, 12248–12253 (2004).
Poggeler, S. & Kuck, U. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378, 1–10 (2006).
Fu, J., Hettler, E. & Wickes, B.L. Split marker transformation increases homologous integration frequency in Cryptococcus neoformans. Fungal Genet. Biol. 43, 200–212 (2006).
Fonzi, W.A. & Irwin, M.Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728 (1993).
Brand, A., MacCallum, D.M., Brown, A.J., Gow, N.A. & Odds, F.C. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3, 900–909 (2004).
Sharkey, L.L., Liao, W.L., Ghosh, A.K. & Fonzi, W.A. Flanking direct repeats of hisG alter URA3 marker expression at the HWP1 locus of Candida albicans. Microbiology 151, 1061–1071 (2005).
Wellington, M., Kabir, M.A. & Rustchenko, E. 5-fluoro-orotic acid induces chromosome alterations in genetically manipulated strains of Candida albicans. Mycologia 98, 393–398 (2006).
Reuss, O., Vik, A., Kolter, R. & Morschhauser, J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127 (2004).
Noble, S.M. & Johnson, A.D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4, 298–309 (2005).
Gerami-Nejad, M., Berman, J. & Gale, C.A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18, 859–864 (2001).
Gerami-Nejad, M., Hausauer, D., McClellan, M., Berman, J. & Gale, C. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast 21, 429–436 (2004).
Gola, S., Martin, R., Walther, A., Dunkler, A. & Wendland, J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20, 1339–1347 (2003).
Walther, A. & Wendland, J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42, 339–343 (2003).
Schaub, Y., Dunkler, A., Walther, A. & Wendland, J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J. Basic Microbiol. 46, 416–429 (2006).
Martin, R., Walther, A. & Wendland, J. Ras1-induced hyphal development in Candida albicans requires the formin Bni1. Eukaryot. Cell 4, 1712–1724 (2005).
Dünkler, A. & Wendland, J. Candida albicans Rho-type GTPase encoding genes required for polarized cell growth and cell separation. Eukaryot. Cell 6, 844–854 (2007).
Martin, R., Hellwig, D., Schaub, Y., Bauer, J., Walther, A. & Wendland, J. Functional analysis of Candida albicans genes whose Saccharomyces cerevisiae homologs are involved in endocytosis. Yeast 24, 511–522 (2007).
Saiki, R.K. et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–1354 (1985).
Roemer, T. et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50, 167–181 (2003).
Kohler, G.A., White, T.C. & Agabian, N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179, 2331–2338 (1997).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989).
Acknowledgements
We thank Hans-Peter Saluz and Grit Mrotzek for providing an introduction into Rapid-PCR. Work in our laboratory is supported by the Deutsche Forschungsgemeinschaft and the European Union—Penelope.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Walther, A., Wendland, J. PCR-based gene targeting in Candida albicans. Nat Protoc 3, 1414–1421 (2008). https://doi.org/10.1038/nprot.2008.137
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.137
This article is cited by
-
Sec15 links bud site selection to polarised cell growth and exocytosis in Candida albicans
Scientific Reports (2016)
-
A new rapid and efficient system with dominant selection developed to inactivate and conditionally express genes in Candida albicans
Current Genetics (2016)
-
One-step targeted gene deletion in Candida albicans haploids
Nature Protocols (2014)
-
Construction of a Kluyveromyces lactis ku80 − Host Strain for Recombinant Protein Production: Extracellular Secretion of Pectin Lyase and a Streptavidin–Pectin Lyase Chimera
Molecular Biotechnology (2014)
-
Candida albicans SH3-domain proteins involved in hyphal growth, cytokinesis, and vacuolar morphology
Current Genetics (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.