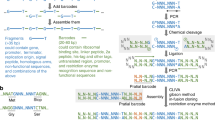
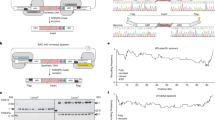
Abstract
A method for synthesizing DNA from 40-mer oligonucleotides, which we used to generate a 32-kb DNA fragment, is explained. DNA sequences are synthesized as ∼500 bp fragments (synthons) in a two-step PCR reaction and cloned using ligation-independent cloning (LIC). Synthons are then assembled into longer full-length sequences in a stepwise manner. By initially synthesizing smaller fragments (synthons), the number of clones sequenced is low compared with synthesizing complete multi-kilobase DNA sequences in a single step. LIC eliminates the need for purification of fragments before cloning, making the process amenable to high-throughput operation and automation. Type IIs restriction enzymes allow seamless assembly of synthons without placing restrictions on the sequence being synthesized. Synthetic fragments are assembled in pairs to generate the final construct using vectors that allow selection of desired clones with two unique antibiotic resistance markers, and this eliminates the need for purification of fragments after digestion with restriction endonucleases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Smith, H.O., Hutchison, C.A., III, Pfannkoch, C. & Venter, J.C. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 100, 15440–15445 (2003).
Dillon, P.J. & Rosen, C.A. A rapid method for the construction of synthetic genes using the polymerase chain reaction. BioTechniques 9, 298–300 (1990).
Prodromou, C. & Pearl, L.H. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 5, 827–829 (1992).
Stemmer, W.P., Crameri, A., Ha, K.D., Brennan, T.M. & Heyneker, H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotide. Gene 164, 49–53 (1995).
Hoover, D.M. & Lubkowski, J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene. Nucleic Acids Res. 30, e43 (2002).
Mandecki, W., Hayden, M.A., Shallcross, M.A. & Stotland, E. A totally synthetic plasmid for general cloning, gene expression and mutagenesis in Escherichia coli. Gene 94, 103–107 (1990).
Cello, J., Paul, A.V. & Wimmer, E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297, 1016–1018 (2002).
Dillon, P.J. & Rosen, C.A. A rapid method for the construction of synthetic genes using the polymerase chain reaction. BioTechniques 9, 298–300 (1990).
Kodumal, S.J. et al. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc. Natl. Acad. Sci. USA 101, 15573–15578 (2004).
Jayaraj, S. et al. GeMS: an advanced software package for designing synthetic genes. Nucleic Acids Res. 33, 3011–3016 (2005).
Hoover, D.M. & Lubkowski, J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 30, e43 (2002).
Rouillard, J.M. et al. Gene2Oligo: oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 32, W176–W180 (2004).
Kodumal, S.J. & Santi, D.V. DNA ligation by selection. BioTechniques 37, 34–40 (2004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Reisinger, S., Patel, K. & Santi, D. Total synthesis of multi-kilobase DNA sequences from oligonucleotides. Nat Protoc 1, 2596–2603 (2006). https://doi.org/10.1038/nprot.2006.426
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.426
This article is cited by
-
Modified ‘one amino acid-one codon’ engineering of high GC content TaqII-coding gene from thermophilic Thermus aquaticus results in radical expression increase
Microbial Cell Factories (2014)
-
Design and testing of a synthetic biology framework for genetic engineering of Corynebacterium glutamicum
Microbial Cell Factories (2012)
-
Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli
Microbial Cell Factories (2011)
-
ICRPfinder: a fast pattern design algorithm for coding sequences and its application in finding potential restriction enzyme recognition sites
BMC Bioinformatics (2009)
-
Preventing the misuse of gene synthesis
Nature Biotechnology (2009)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.