Abstract
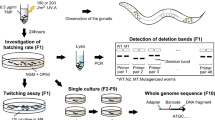
This protocol details methodologies to generate Caenorhabditis elegans deletion mutants by chemical mutagenesis and to detect them by PCR screening. Approximately, 600,000 worms are grown synchronously, mutagenized with ethyl methane sulfonate, divided in groups of 500 and allowed to self-fertilize for two generations. DNA is prepared from a fraction of each worm population, pooled into a 96-well plate, and screened by PCR with primers positioned 2.5–3.5 kb apart. Cultures containing deletion mutants are subdivided in small worm populations and tested again by PCR to identify positives. Single animals are then cloned from positive cultures, allowed to self-fertilize and identified by PCR genotyping. This method, which takes about a month, gives approximately a 50% chance of finding a deletion of interest larger than 500–600 bp. If a deletion cannot be found, the library can be pooled at lower complexity and screened for smaller deletions using an alternative PCR-based method.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
29 December 2006
In the version of this article initially published online the background colors for Figure 1 were omitted. This error has been corrected in all versions of the article.
References
Zwaal, R.R., Broeks, A., van Meurs, J., Groenen, J.T.M. & Plasterk, R.H.A. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl. Acad. Sci. USA 90, 7431–7435 (1993).
Bessereau, J.L. et al. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413, 70–74 (2001).
Jansen, G., Hazendonk, E., Thijssen, K.L. & Plasterk, R.H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 17, 119–121 (1997).
Anderson, P. Mutagenesis. Methods Cell Biol. 48, 31–58 (1995).
Gengyo-Ando, K. & Mitani, S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269, 64–69 (2000).
Yandell, M.D., Edgar, L.G. & Wood, W.B. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 91, 1381–1385 (1994).
Liu, L.X. et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 9, 859–867 (1999).
Shen, X. et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893–903 (2001).
Lesa, G.M. et al. Long chain polyunsaturated fatty acids are required for efficient neurotransmission in C. elegans. J. Cell Sci. 116, 4965–4975 (2003).
Bui, Y.K. & Sternberg, P.W. Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol. Biol. Cell 13, 1641–1651 (2002).
Barstead, R.J. Reverse genetics in C. elegans. in A Practical Approach (ed. Hope, I.A.) (Oxford University Press, Oxford, 1999).
Hess, H., Reinke, V. & Koelle, M. Construction and Screening of Deletion Mutant Libraries to Generate C. elegans Gene Knockouts in Wormbook. (ed. Ahringer, J.) (The C. elegans Research Community, 2006).
Edgley, M. et al. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 30, e52 (2002).
In the version of this article initially published online the background colors for Figure 1 were omitted. This error has been corrected in all versions of the article.
Acknowledgements
I thank G Schiavo for help in setting up the protocol and G Lalli and N Hopper for suggestions. Work in my laboratory was funded by the Royal Society and by the Medical Research Council. Part of the work described here was funded by Cancer Research UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary Data 1
Customizable PCR program for Primary and Secondary PCRs (XLS 24 kb)
Supplementary Data 2
Customizable PCR program for Genotyping PCR (XLS 20 kb)
Rights and permissions
About this article
Cite this article
Lesa, G. Isolation of Caenorhabditis elegans gene knockouts by PCR screening of chemically mutagenized libraries. Nat Protoc 1, 2231–2240 (2006). https://doi.org/10.1038/nprot.2006.345
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.345
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.