Abstract
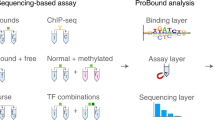
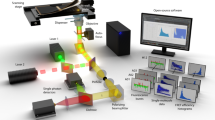
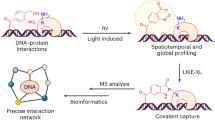
Limited information exists for the binding specificities of many important transcription factors. To address this, we have previously developed a microwell-based assay for directly measuring the affinity of DNA-protein binding interactions. We describe here the detailed protocol for determining sequence specificities of DNA-binding proteins using this assay. The described method is rapid; after preparation of the reagents, the assay can be run in a single day, and its throughput can be increased further by automation. The method is quantitative but requires prior knowledge of one high-affinity binding site for the protein of interest. The protocol can be adapted for determining the effect of protein modifications and protein-protein interactions on DNA-binding specificity, and for engineering proteins with new DNA-binding specificities. In addition, the method is suitable for high-throughput screening to identify proteins or small molecules that modulate protein-DNA binding interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Eisenstein, M. Search party. Nat. Methods 3, 153 (2006).
Holstege, F.C. & Clevers, H. Transcription factor target practice. Cell 124, 21–23 (2006).
Hallikas, O. et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124, 47–59 (2006).
Roulet, E. et al. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 20, 831–835 (2002).
Mukherjee, S. et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 36, 1331–1339 (2004).
Warren, C.L. et al. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl Acad. Sci. USA 103, 867–872 (2006).
Sambrook, J. & Russell, D.W. in Molecular Cloning: a Laboratory Manual 3rd edn., Vol. 3 17.4–17.12 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Kinzler, K.W. & Vogelstein, B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 10, 634–642 (1990).
Ptashne, M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 30, 275–279 (2005).
Passner, J.M., Ryoo, H.D., Shen, L., Mann, R.S. & Aggarwal, A.K. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397, 714–719 (1999).
Panne, D., Maniatis, T. & Harrison, S.C. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 23, 4384–4393 (2004).
Golemis, E.A. Protein interaction technologies. In Molecular Cloning: a Laboratory Manual 3rd edn., Vol. 3 (eds. Sambrook, J & Russell, D.W.) 18.1–18.36 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Wei, C.L. et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 (2006).
Motulsky, H. & Christopoulos, A. In Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting 211–232 (Oxford University Press, New York, 2004).
Acknowledgements
We thank H. Clevers for constructs and M. Björklund and A. Oinas for reviewing the protocol. This work was supported by the Translational Genome-Scale Biology Center of Excellence of the Academy of Finland, the Regulatory Genomics specific targeted research project of the European Union, Biocentrum Helsinki, the University of Helsinki, the Sigrid Juselius Foundation, the Magnus Ehrnrooth Foundation, the Jenny and Antti Wihuri Foundation and the Finnish Cancer Research Organizations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hallikas, O., Taipale, J. High-throughput assay for determining specificity and affinity of protein-DNA binding interactions. Nat Protoc 1, 215–222 (2006). https://doi.org/10.1038/nprot.2006.33
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.33
This article is cited by
-
Differentiation capacity of dental pulp stem cell into inner ear hair cell using an in vitro assay: a preliminary step toward treating sensorineural hearing loss
European Archives of Oto-Rhino-Laryngology (2022)
-
Insights gained from a comprehensive all-against-all transcription factor binding motif benchmarking study
Genome Biology (2020)
-
Non-consensus GLI binding sites in Hedgehog target gene regulation
BMC Molecular Biology (2010)
-
Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo
The EMBO Journal (2010)
-
The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling
Nature Genetics (2009)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.