Abstract
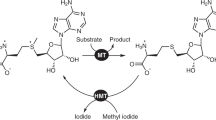
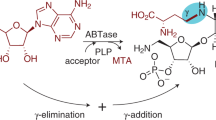
Here we describe a one-step synthetic procedure for the preparation of S-adenosyl-L-methionine (AdoMet) analogs with extended carbon chains replacing the methyl group. These AdoMet analogs function as efficient cofactors for DNA methyltransferases (MTases), and we provide a protocol for sequence-specific transfer of extended side chains from these AdoMet analogs to DNA by DNA MTases. Direct chemoselective allylation or propargylation of S-adenosyl-L-homocysteine (AdoHcy) at sulfur is achieved under the acidic conditions needed to protect other nucleophilic positions in AdoHcy. The unsaturated bonds in β position to the sulfonium center of the resulting AdoMet analogs are designed to stabilize the transition state formed upon DNA MTase-catalyzed nucleophilic attack at the carbon next to the sulfonium center and lead to efficient transfer of the extended side chains to DNA. Using these protocols, sequence-specific functionalized DNA can be obtained within one to two weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chiang, P.K. et al. S-Adenosylmethionine and methylation. FASEB J. 10, 471–480 (1996).
Cheng, X. & Blumenthal, R.M. S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions 392 (World Scientific, Singapore, 1999).
Parks, L.W. S-Adenosylethionine and ethionine inhibition. J. Biol. Chem. 232, 169–176 (1958).
Schlenk, F. & Dainko, J.L. The S-n-propyl analogue of S-adenosylmethionine. Biochim. Biophys. Acta 385, 312–323 (1975).
Schlenk, F. Recent studies of the chemical properties of adenosylmethionine and related compounds. In The Biochemistry of Adenosylmethionine (eds. Salvatore, F., Borek, E. Zappia, V., Williams-Ashman, H.G. & Schlenk, F.) 3–17 (Columbia University Press, New York, 1977).
Dalhoff, C., Lukinavičius, G., Klimašauskas, S. & Weinhold, E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat. Chem. Biol. 2, 31–32 (2006).
Cantoni, G.L. The nature of the active methyl donor formed enzymatically from L-methionine and adenosinetriphosphate. J. Am. Chem. Soc. 74, 2942–2943 (1952).
De La Haba, G., Jamieson, G.A., Mudd, S.H. & Richards, H.H. S-Adenosylmethionine: the relation of configuration at the sulfonium center to enzymatic reactivity. J. Am. Chem. Soc. 81, 3975–3980 (1959).
Ross, S.A., Pitié, M. & Meunier, B. A straightforward preparation of primary alkyl triflates and their utility in the synthesis of derivatives of ethidium. J. Chem. Soc. Perkin Trans. 1, 571–574 (2000).
Borchardt, R.T. & Wu, Y.S. Potential inhibitors of S-adenosylmethionine-dependent methyltransferases. 5. Role of the asymmetric sulfonium pole in the enzymatic binding of S-adenosyl-L-methionine. J. Med. Chem. 19, 1099–1103 (1976).
Gupta, V. & Kool, E.T. A self-cleaving DNA nucleoside. Chem. Commun. 1425–1426 (1997).
Kubo, K., Ide, H., Wallace, S.S. & Kow, Y.W.A. Novel, sensitive, and specific assay for abasic sites, the most commonly produced DNA lesion. Biochemistry 31, 3703–3708 (1992).
Pljevaljcic, G., Schmidt, F. & Weinhold, E. Sequence-specific methyltransferase-induced labeling of DNA (SMILing DNA). ChemBioChem. 5, 265–269 (2004).
Holz, B., Klimasauskas, S., Serva, S. & Weinhold, E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 26, 1076–1083 (1998).
Goedecke, K., Pignot, M., Goody, R.S., Scheidig, A.J. & Weinhold, E. Structure of the N6-adenine DNA methyltransferase M·TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 8, 121–125 (2001).
Cantor, C.R., Warshaw, M.M. & Shapiro, H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers 9, 1059–1077 (1970).
Connolly, B.A. Synthetic oligodeoxynucleotides containing modified bases. Methods Enzymol. 211, 36–53 (1992).
Jones, J.W. & Robins, R.K. Purine nucleosides. III. Methylation studies of certain naturally occurring purine nucleosides. J. Am. Chem. Soc. 85, 193–201 (1963).
Sambrook, J. & Russel, D.W. Molecular Cloning: A Laboratory Manual 3rd edn. Vol. 1, 5.4–5.13 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2001).
Acknowledgements
The authors are grateful to K. Glensk for preparing M.TaqI. C.D. thanks the Deutsche Forschungsgemeinschaft for a stipend within the Graduiertenkolleg 440. G.L. thanks M. Krenevièienë for recording NMR spectra for cofactor analog 4. The M.TaqI expression vector pAGL15-M13 was kindly provided by J. Benner and C. Baxa, New England Biolabs. This work was supported by grants from the VolkswagenStiftung, the Howard Hughes Medical Institute and the Lithuanian Science and Study Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
We have filed a patent application on the doubly activated AdoMet analogs and made a license agreement with a company. Thus, we are eligible for possible future personal royalties.
Rights and permissions
About this article
Cite this article
Dalhoff, C., Lukinavičius, G., Klimašauskas, S. et al. Synthesis of S-adenosyl-L-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases. Nat Protoc 1, 1879–1886 (2006). https://doi.org/10.1038/nprot.2006.253
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.253
This article is cited by
-
Methods for detection of cytosine and thymine modifications in DNA
Nature Reviews Chemistry (2018)
-
Refining the pathway of carbide insertion into the nitrogenase M-cluster
Nature Communications (2015)
-
Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.