Abstract
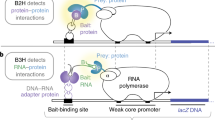
To test RNA-RNA interactions in cells, we developed a yeast RNA hybrid system derived from the yeast three-hybrid system. In this setup, the activation of a reporter gene (HIS3 or lacZ) is dependent on the interaction of two RNAs. One ('RNA X') is fused to MS2 RNA, forming the bait, which binds to a fusion protein composed of the MS2 coat and the LexA proteins. The second ('RNA Y') is fused to an RNA-based transcriptional activator (m26-11), forming the prey. If prey (RNA Y) binds to bait (RNA X), the m26-11 transcriptional activator is tethered to the promoter of the reporter genes. This protocol describes how to use this RNA hybrid system. In addition to testing RNA-RNA interactions, it can also be used to screen RNA libraries to identify new interaction partners or for mutational analysis of two known interaction partners.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gottesman, S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58, 303–328 (2004).
Tucker, B.J. & Breaker, R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 15, 342–348 (2005).
Wagner, E.G. & Simons, R.R. Antisense RNA control in bacteria, phage and plasmids. Annu. Rev. Microbiol. 48, 713–742 (1994).
SenGupta, D.J. et al. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 93, 8496–501 (1996).
Fields, S. & Song, O. A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 (1989).
Buskirk, A.R., Kehayova, P.D., Landrigan, A. & Liu, D.R. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 10, 533–540 (2003).
Buskirk, A.R., Landrigan, A. & Liu, D.R. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 11, 1157–1163 (2004).
Buskirk, A.R., Ong, Y.C., Gartner, Z.J. & Liu, D.R. Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. USA 101, 10505–10510 (2004).
Piganeau, N., Schauer, U.E. & Schroeder, R. A yeast RNA-hybrid system for the detection of RNA-RNA interactions in vivo. RNA 12, 177–184 (2005).
Zucker, M. Mfold web server for nucleic acids folding and hybridization prediction. Nucl. Acids Res. 31, 3406–3415 (2003).
Hook, B., Bernstein, D., Zhang, B. & Wickens, M. RNA-protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA 11, 227–233 (2005).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular Cloning: a Laboratory Manual, A2.9–A2.11 (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1989).
Inoue, H., Nojima, H. & Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 96, 23–28 (1990).
Schiestl, R.H. & Gietz, R.D. High efficiency transformation on intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16, 339–346 (1989).
Bernstein, D.S., Buter, N., Stumpf, C. & Wickens, M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26, 123–141 (2002).
Lorenz, C., Piganeau, N. & Schroeder, R. Stabilities of HIV-1 DIS type RNA loop-loop interactions in vitro and in vivo. Nucl. Acids Res. 34, 334–342 (2006).
Acknowledgements
Work in our laboratory is funded by the Austrian Science Fund (projects Z-72 and F1703), by the European community Research Programme Framework 6 (BACRNA FP6-018618) and by the Austrian BMBWK GenAU program.
Author information
Authors and Affiliations
Contributions
N.P. developed the method and wrote most of this experimental procedure; R.S. is the supervisor and wrote the introduction and the anticipated results sections of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
pIIIa m26 and pAN MS2-2. (PDF 25 kb)
Supplementary Data 1
pAN MS2-2. (TXT 9 kb)
Supplementary Data 2
pIIIa m26. (TXT 11 kb)
Supplementary Data 3
pAN X1 MS2. (TXT 9 kb)
Supplementary Data 4
pIIIa m26 Y1. (TXT 11 kb)
Rights and permissions
About this article
Cite this article
Piganeau, N., Schroeder, R. Identification and detection of RNA-RNA interactions using the yeast RNA hybrid system. Nat Protoc 1, 689–694 (2006). https://doi.org/10.1038/nprot.2006.111
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.111
This article is cited by
-
Advances and challenges towards the study of RNA-RNA interactions in a transcriptome-wide scale
Quantitative Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.