Abstract
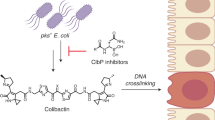
Various forms of cancer have been linked to the carcinogenic activities of microorganisms1–3. The virulent gene island polyketide synthase (pks) produces the secondary metabolite colibactin, a genotoxic molecule(s) causing double-stranded DNA breaks4 and enhanced colorectal cancer development5,6. Colibactin biosynthesis involves a prodrug resistance strategy where an N-terminal prodrug scaffold (precolibactin) is assembled, transported into the periplasm and cleaved to release the mature product7–10. Here, we show that ClbM, a multidrug and toxic compound extrusion (MATE) transporter, is a key component involved in colibactin activity and transport. Disruption of clbM attenuated pks+ E. coli-induced DNA damage in vitro and significantly decreased the DNA damage response in gnotobiotic Il10−/− mice. Colonization experiments performed in mice or zebrafish animal models indicate that clbM is not implicated in E. coli niche establishment. The X-ray structure of ClbM shows a structural motif common to the recently described MATE family. The 12-transmembrane ClbM is characterized as a cation-coupled antiporter, and residues important to the cation-binding site are identified. Our data identify ClbM as a precolibactin transporter and provide the first structure of a MATE transporter with a defined and specific biological function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jobin, C. Colorectal cancer: looking for answers in the microbiota. Cancer Discov. 3, 384–387 (2013).
Schwabe, R. F. & Jobin, C. The microbiome and cancer. Nature Rev. Cancer 13, 800–812 (2013).
Sears, C. L. & Garrett, W. S. Microbes, microbiota, and colon cancer. Cell Host Microbe 15, 317–328 (2014).
Nougayrède, J.-P. et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006).
Arthur, J. C. et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nature Commun. 5, 4724 (2014).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Dubois, D. et al. ClbP is a prototype of a peptidase subgroup involved in biosynthesis of nonribosomal peptides. J. Biol. Chem. 286, 35562–35570 (2011).
Cougnoux, A. et al. Analysis of structure–function relationships in the colibactin-maturating enzyme ClbP. J. Mol. Biol. 424, 203–214 (2012).
Brotherton, C. A. & Balskus, E. P. A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J. Am. Chem. Soc. 135, 3359–3362 (2013).
Bian, X. et al. In vivo evidence for a prodrug activation mechanism during colibactin maturation. ChemBioChem 14, 1194–1197 (2013).
Buc, E. et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 8, e56964 (2013).
Vizcaino, M. I., Engel, P., Trautman, E. & Crawford, J. M. Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J. Am. Chem. Soc. 136, 9244–9247 (2014).
Brotherton, C. A., Wilson, M., Byrd, G. & Balskus, E. P. Isolation of a metabolite from the pks island provides insights into colibactin biosynthesis and activity. Org. Lett. 17, 1545–1548 (2015).
Bian, X., Plaza, A., Zhang, Y. & Mueller, R. Two more pieces of the colibactin genotoxin puzzle from Escherichia coli show incorporation of an unusual 1-aminocyclopropanecarboxylic acid moiety. Chem. Sci. 3154–3160 (2015).
Vizcaino, M. I. & Crawford, J. M. The colibactin warhead crosslinks DNA. Nature Chem. 7, 411–417 (2015).
Li, Z.-R. et al. Critical intermediates reveal new biosynthetic events in the enigmatic colibactin pathway. ChemBioChem 16, 1715–1719 (2015).
Brachmann, A. O. et al. Colibactin biosynthesis and biological activity depend on the rare aminomalonyl polyketide precursor. Chem. Commun. 3034, 13138–13141 (2015).
Kaatz, G. W., Mcaleese, F. & Seo, S. M. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49, 1857–1864 (2005).
Mcaleese, F. et al. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother. 49, 1865–1871 (2005).
Long, F., Rouquette-Loughlin, C., Shafer, W. M. & Yu, E. W. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli. Antimicrob. Agents Chemother. 52, 3052–3060 (2008).
Kuroda, T. & Tsuchiya, T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794, 763–768 (2009).
Nikaido, H. & Zgurskaya, H. I. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3, 215–218 (2001).
Kinner, A., Wu, W., Staudt, C. & Iliakis, G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36, 5678–5694 (2008).
Cherezov, V. Lipidic cubic phase technologies for membrane protein structural studies. Curr. Opin. Struct. Biol. 21, 559–566 (2011).
Von Heijne, G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494 (1992).
He, X. et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994 (2010).
Lu, M. et al. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl Acad. Sci. USA 110, 2099–2104 (2013).
Lu, M., Radchenko, M., Symersky, J., Nie, R. & Guo, Y. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nature Struct. Mol. Biol. 20, 1310–1317 (2013).
Tanaka, Y. et al. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496, 247–251 (2013).
Li, Z. et al. Critical intermediates reveal novel biosynthetic events in the enigmatic colibactin pathway. ChemBioChem 16, 1715–1719 (2015).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Martin, P. et al. Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog. 9, e1003437 (2013).
Huang, X. & Darzynkiewicz, Z. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol. Bio. 314, 73–80 (2006).
Semova, I. et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288 (2012).
Chen, J. et al. VmrA, a member of a novel class of Na+-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J. Bacteriol. 184, 572–576 (2002).
Morita, Y. et al. NorM, a putative multidrug efflux protein, of vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782 (1998).
Filip, C., Fletcher, G., Wulff, J. L. & Earhart, C. F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115, 717–722 (1973).
Miroux, B. & Walker, J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
This work was supported by funding from R01 DK73338 (C.J.), R01DK47700 (C.J.), R01 GM0865700 (S.D.B.) and by the University of Florida (S.D.B.) and the UF DoM Gatorade Fund (C.J.). The authors thank V. de Crecy-Lagard for discussions.
Author information
Authors and Affiliations
Contributions
C.J., S.D.B. and E.O. conceived the project and designed the experiments. J.J.M., Y.Y., S.D.B. and C.J. wrote the manuscript. J.J.M. purified, crystallized, collected data and solved the structure of ClbM. J.J.M. and R.C.N. performed the ethidium fluorescence assay and LC-MS analysis of compound 1 accumulation. J.J.M., P.T. and Y.Y. performed the ClbM cellular localization experiment. Y.Y. made ΔclbM and ΔacrA in NC101 and performed the in vitro infection and mouse colonization studies. S.T. carried out the zebrafish colonization study. A.S. constructed MGpks+ and MGpks+ mutants and examined minimal inhibitory concentrations of various antibiotics and toxic compounds. Y.Y. performed statistical analyses. J.J.M. and Y.Y. contributed equally to this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–9 and Tables 1,2. (PDF 1329 kb)
Supplementary Video 1
NC101-colonized, fixed 6-d.p.f. zebrafish. Reconstruction from z-stack images; DAPI stained whole mount zebrafish with tdTomato-labelled bacteria. (MOV 1444 kb)
Supplementary Video 2
ΔclbM-colonized, fixed 6-d.p.f. zebrafish. Reconstruction from z-stack images; DAPI stained whole mount zebrafish with tdTomato-labelled bacteria. (MOV 1559 kb)
Rights and permissions
About this article
Cite this article
Mousa, J., Yang, Y., Tomkovich, S. et al. MATE transport of the E. coli-derived genotoxin colibactin. Nat Microbiol 1, 15009 (2016). https://doi.org/10.1038/nmicrobiol.2015.9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2015.9
This article is cited by
-
Engineered MATE multidrug transporters reveal two functionally distinct ion-coupling pathways in NorM from Vibrio cholerae
Communications Biology (2021)
-
“Effect of Subinhibitory Concentrations of Some Antibiotics and Low Doses of Gamma Radiation on the Cytotoxicity and Expression of Colibactin by an Uropathogenic Escherichia coli isolate”
Current Microbiology (2021)
-
Prevalence and pathologic effects of colibactin and cytotoxic necrotizing factor-1 (Cnf 1) in Escherichia coli: experimental and bioinformatics analyses
Gut Pathogens (2019)
-
Microbial Colonization Coordinates the Pathogenesis of a Klebsiella pneumoniae Infant Isolate
Scientific Reports (2019)
-
Multidrug efflux pumps: structure, function and regulation
Nature Reviews Microbiology (2018)