Abstract
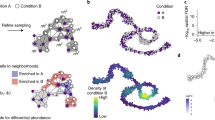
When comparing biological conditions using mass cytometry data, a key challenge is to identify cellular populations that change in abundance. Here, we present a computational strategy for detecting 'differentially abundant' populations by assigning cells to hyperspheres, testing for significant differences between conditions and controlling the spatial false discovery rate. Our method (http://bioconductor.org/packages/cydar) outperforms other approaches in simulations and finds novel patterns of differential abundance in real data.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ornatsky, O.I. et al. Anal. Chem. 80, 2539–2547 (2008).
Leipold, M.D., Newell, E.W. & Maecker, H.T. Methods Mol. Biol. 1343, 81–95 (2015).
Leelatian, N., Diggins, K.E. & Irish, J.M. Methods Mol. Biol. 1346, 99–113 (2015).
Hansmann, L. et al. Cancer Immunol. Res. 3, 650–660 (2015).
Bendall, S.C. et al. Science 332, 687–696 (2011).
Levine, J.H. et al. Cell 162, 184–197 (2015).
Qiu, P. et al. Nat. Biotechnol. 29, 886–891 (2011).
Samusik, N., Good, Z., Spitzer, M.H., Davis, K.L. & Nolan, G.P. Nat. Methods 13, 493–496 (2016).
Gaudillière, B. et al. Sci. Transl. Med. 6, 255ra131 (2014).
Gaudillière, B. et al. Cytometry A 87, 817–829 (2015).
Anchang, B. et al. Nat. Protoc. 11, 1264–1279 (2016).
Bruggner, R.V., Bodenmiller, B., Dill, D.L., Tibshirani, R.J. & Nolan, G.P. Proc. Natl. Acad. Sci. USA 111, E2770–E2777 (2014).
Ronan, T., Qi, Z. & Naegle, K.M. Sci. Signal. 9, re6 (2016).
Finak, G. et al. Sci. Rep. 6, 20686 (2016).
McCarthy, D.J., Chen, Y. & Smyth, G.K. Nucleic Acids Res. 40, 4288–4297 (2012).
Benjamini, Y. & Hochberg, Y. Scand. J. Stat. 24, 407–418 (1997).
Zunder, E.R., Lujan, E., Goltsev, Y., Wernig, M. & Nolan, G.P. Cell Stem Cell 16, 323–337 (2015).
Van der Maaten, L. & Hinton, G. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Parks, D.R., Roederer, M. & Moore, W.A. Cytometry A 69, 541–551 (2006).
Hahne, F. et al. BMC Bioinformatics 10, 106 (2009).
Acknowledgements
This work was supported by Cancer Research UK (core funding to J.C.M., award no. A17197), the University of Cambridge and Hutchison Whampoa Limited. J.C.M. was also supported by core funding from EMBL.
Author information
Authors and Affiliations
Contributions
A.T.L.L. developed the analysis pipeline, tested it with simulations and applied it to the real data. A.C.R. interpreted the results to identify the DA subpopulations. J.C.M. provided direction and advice on method development and biological interpretation. All authors wrote and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–24, Supplementary Table 1 and Supplementary Notes 1–9. (PDF 15317 kb)
Supplementary Software
cydar software. (ZIP 190 kb)
Source data
Rights and permissions
About this article
Cite this article
Lun, A., Richard, A. & Marioni, J. Testing for differential abundance in mass cytometry data. Nat Methods 14, 707–709 (2017). https://doi.org/10.1038/nmeth.4295
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4295
This article is cited by
-
Benchmarking differential abundance methods for finding condition-specific prototypical cells in multi-sample single-cell datasets
Genome Biology (2024)
-
Single-cell insights into immune dysregulation in rheumatoid arthritis flare versus drug-free remission
Nature Communications (2024)
-
Semi-supervised integration of single-cell transcriptomics data
Nature Communications (2024)
-
Trajectory inference across multiple conditions with condiments
Nature Communications (2024)
-
A comparative study of in vitro air–liquid interface culture models of the human airway epithelium evaluating cellular heterogeneity and gene expression at single cell resolution
Respiratory Research (2023)