Abstract
Molecular biomarkers can serve as useful diagnostic markers, as prognostic markers for predicting clinical behavior, or as targets for new therapeutic strategies. Application of expression microarray technology, which allows the expression of all or most of the genes in the human genome to be analyzed simultaneously, has dramatically enhanced the discovery of prostate cancer biomarkers. The diagnostic markers identified include AMACR (α-methylacyl CoA racemase), a protein that has already been translated into clinical use as an aid in distinguishing prostate cancer from benign disease. Individual genes, such as the polycomb gene EZH2 whose expression indicates poor survival, have been identified. The power of microarray technology is that it has allowed the identification of gene signatures (each composed of multiple genes) that might provide improved prediction of clinical outcomes in human prostate cancer. The development of a new method for analyzing expression microarray data, called COPA, has led to the discovery of TMPRSS2–ERG gene fusion involvement in the development of prostate cancer, while expression analysis of castration-resistant prostate cancer has suggested the use of novel therapeutic approaches for advanced disease. Despite these successes, there are limitations in the application of microarray technology to prostate cancer; for example, unlike with other cancers, this approach has failed to provide a consistent unsupervised classification of the disease. Overcoming the reasons for these failures represents a major challenge for future research endeavors.
Key Points
-
Analyses of expression microarray data have resulted in the identification of many individual diagnostic and prognostic markers, including AMACR, EZH2, AZGP1, MEMD/CD166 and CD24
-
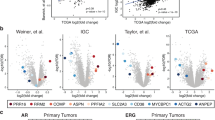
Cancer outlier profile analysis of expression microarray data demonstrated that ERG and ETV1 genes are overexpressed in a subset of cancers, leading to the discovery that these genes are activated by the formation of TMPRSS2–ERG and TMPRSS2–ETV1 gene fusions
-
Observations from microarray expression studies that the androgen receptor pathway is still activated in castration-resistant prostate cancer (CRPC) has led to the discovery that total androgen blockade with abiraterone might be an effective treatment for CRPC
-
Published expression microarray datasets are relatively small (maximum 79 prostate cancers); there is an urgent need to collect larger datasets that take issues such as genetic heterogeneity and cancer multifocality into consideration during the preparation of RNA samples used in microarray studies
-
Elucidation of the role of microRNAs in the development and clinical management of prostate cancer represents an important goal for future research
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Quinn DI et al. (2005) Molecular markers of prostate cancer outcome. Eur J Cancer 41: 858–887
Jemal A et al. (2007) Cancer statistics, 2007. CA Cancer J Clin 57: 43–66
Yao SL et al. (2002) Understanding and appreciating overdiagnosis in the PSA era. J Natl Cancer Inst 94: 958–960
Glinsky GV et al. (2004) Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest 113: 913–923
Yu YP et al. (2004) Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 22: 2790–2799
Djavan B et al. (1999) Predictability and significance of multifocal prostate cancer in the radical prostatectomy specimen. Tech Urol 5: 139–142
Arora R et al. (2004) Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer 100: 2362–2366
Chen ME et al. (2000) Detailed mapping of prostate carcinoma foci: biopsy strategy implications. Cancer 89: 1800–1809
Miller GJ and Cygan JM (1994) Morphology of prostate cancer: the effects of multifocality on histological grade, tumor volume and capsule penetration. J Urol 152: 1709–1713
Villers A et al. (1992) Multiple cancers in the prostate. Morphologic features of clinically recognized versus incidental tumors. Cancer 70: 2313–2318
Aihara M et al. (1994) Heterogeneity of prostate cancer in radical prostatectomy specimens. Urology 43: 60–66
Bostwick DG et al. (1998) Independent origin of multiple foci of prostatic intraepithelial neoplasia: comparison with matched foci of prostate carcinoma. Cancer 83: 1995–2002
Cheng L et al. (1998) Evidence of independent origin of multiple tumors from patients with prostate cancer. J Natl Cancer Inst 90: 233–237
Qian J et al. (1995) Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Res 55: 5408–5414
Jenkins RB et al. (1997) Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 57: 524–531
Konishi N et al. (1995) Intratumor cellular heterogeneity and alterations in ras oncogene and p53 tumor suppressor gene in human prostate carcinoma. Am J Pathol 147: 1112–1122
Mirchandani D et al. (1995) Heterogeneity in intratumor distribution of p53 mutations in human prostate cancer. Am J Pathol 147: 92–101
Rhodes DR et al. (2002) Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res 62: 4427–4433
Golub TR et al. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537
Alizadeh AA et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511
Sorlie T et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874
Sorlie T et al. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423
Lapointe J et al. (2004) Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA 101: 811–816
Dhanasekaran SM et al. (2001) Delineation of prognostic biomarkers in prostate cancer. Nature 412: 822–826
Rogers S et al. (2005) The latent process decomposition of cDNA microarray data sets. IEEE/ACM Trans Comput Biol Bioinform 2: 143–156
Rogers S and Girolami M (2005) A Bayesian regression approach to the inference of regulatory networks from gene expression data. Bioinformatics 21: 3131–3137
Carrivick L et al. (2006) Identification of prognostic signatures in breast cancer microarray data using Bayesian techniques. J R Soc Interface 3: 367–381
Xu J et al. (2001) Identification and characterization of prostein, a novel prostate-specific protein. Cancer Res 61: 1563–1568
Liu XF et al. (2001) PRAC: A novel small nuclear protein that is specifically expressed in human prostate and colon. Prostate 47: 125–131
Olsson P et al. (2003) PRAC2: a new gene expressed in human prostate and prostate cancer. Prostate 56: 123–130
Edwards S et al. (2005) Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Br J Cancer 92: 376–381
Hubert RS et al. (1999) STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci USA 96: 14523–14528
Porkka KP et al. (2002) Cloning and characterization of a novel six-transmembrane protein STEAP2, expressed in normal and malignant prostate. Lab Invest 82: 1573–1582
Korkmaz KS et al. (2002) Molecular cloning and characterization of STAMP1, a highly prostate-specific six transmembrane protein that is overexpressed in prostate cancer. J Biol Chem 277: 36689–36696
Korkmaz CG et al. (2005) Molecular cloning and characterization of STAMP2, an androgen-regulated six transmembrane protein that is overexpressed in prostate cancer. Oncogene 24: 4934–4945
Varambally S et al. (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629
Bracken AP et al. (2003) EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22: 5323–5335
Bryant RJ et al. (2007) EZH2 promotes proliferation and invasiveness of prostate cancer cells. Prostate 67: 547–556
Rubin MA et al. (2002) α-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA 287: 1662–1670
Rubin MA et al. (2005) Decreased α-methylacyl CoA racemase expression in localized prostate cancer is associated with an increased rate of biochemical recurrence and cancer-specific death. Cancer Epidemiol Biomarkers Prev 14: 1424–1432
Luo J et al. (2001) Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res 61: 4683–4688
Welsh JB et al. (2001) Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res 61: 5974–5978
Magee JA et al. (2001) Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res 61: 5692–5696
Klezovitch O et al. (2004) Hepsin promotes prostate cancer progression and metastasis. Cancer Cell 6: 185–195
Kristiansen G et al. (2005) Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. J Pathol 205: 359–376
Stephenson AJ et al. (2005) Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer 104: 290–298
Bismar TA et al. (2006) Defining aggressive prostate cancer using a 12-gene model. Neoplasia 8: 59–68
Bibikova M et al. (2007) Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics 89: 666–672
Ein-Dor L et al. (2006) Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA 103: 5923–5928
Tomlins SA et al. (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648
Demichelis F et al. (2007) TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene 26: 4596–4599
Wang J et al. (2006) Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res 66: 8347–8351
Nami RK et al. (2007) Expression of TMPRSS2 ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther 6: 40–45
Perner S et al. (2006) TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res 66: 8337–8341
Attard G et al. (2007) Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene [10.1038/sj.onc.1210640]
Takayama K et al. (2007) Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene 26: 4453–4463
Chen CD et al. (2004) Molecular determinants of resistance to antiandrogen therapy. Nat Med 10: 33–39
Holzbeierlein J et al. (2004) Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol 164: 217–227
Attard G et al. (2005) Selective blockade of androgenic steroid synthesis by novel lyase inhibitors as a therapeutic strategy for treating metastatic prostate cancer. BJU Int 96: 1241–1246
de Bono J. et al. (2006) Inhibition of androgen synthesis by an oral, irreversible, inhibitor of CYP450c17 is safe and results in a high, durable, response rate in castration refractory prostate cancer (CRPC) patients [abstract]. Presented at the NCRI Cancer Conference: 2006 October 8–11, Birmingham, UK
Ramaswamy S et al. (2003) A molecular signature of metastasis in primary solid tumors. Nat Genet 33: 49–54
Carter SL et al. (2006) A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 38: 1043–1048
Glinsky GV et al. (2005) Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 115: 1503–1521
Lahad JP et al. (2005) Stem cell-ness: a “magic marker” for cancer. J Clin Invest 115: 1463–1467
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297
Lee RC et al. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell 75: 843–854
Chan JA et al. (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033
Croce CM and Calin GA (2005) miRNAs, cancer, and stem cell division. Cell 122: 6–7
Lu J et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838
Volinia S et al. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261
Porkka KP et al. (2007) MicroRNA expression profiling in prostate cancer. Cancer Res 67: 6130–6135
Chiosea S et al. (2006) Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol 169: 1812–1820
Jhavar SG et al. (2005) Processing of radical prostatectomy specimens for correlation of data from histopathological, molecular biological, and radiological studies: a new whole organ technique. J Clin Pathol 58: 504–508
Iljin K et al. (2006) TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res 66: 10242–10246
Singh D et al. (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1: 203–209
Chandran UR et al. (2005) Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer 5: 45
Halvorsen OJ et al. (2005) Gene expression profiles in prostate cancer: association with patient subgroups and tumour differentiation. Int J Oncol 26: 329–336
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Cooper, C., Campbell, C. & Jhavar, S. Mechanisms of Disease: biomarkers and molecular targets from microarray gene expression studies in prostate cancer. Nat Rev Urol 4, 677–687 (2007). https://doi.org/10.1038/ncpuro0946
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ncpuro0946
This article is cited by
-
Integrative Analysis of Potential Biomarkers Involved in the Progression of Papillary Thyroid Cancer
Applied Biochemistry and Biotechnology (2023)
-
From transcriptome analysis to therapeutic anti-CD40L treatment in the SOD1 model of amyotrophic lateral sclerosis
Nature Genetics (2010)
-
Innovations in the systemic therapy of prostate cancer
Nature Reviews Clinical Oncology (2010)
-
Differential expression of apoptotic genes PDIA3 and MAP3K5 distinguishes between low- and high-risk prostate cancer
Molecular Cancer (2009)