Abstract
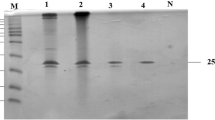
The Streptomyces lividans xylanase A (XlnA) signal peptide (sp) was replaced with the signal peptides of either mannanase A (ManA) or cellulase A (CelA), two enzymes secreted by S. lividans. Depending on the location of the ribosome binding sites (RBS) with respect to a potential initiation codon, the length of the putative sps of ManA and CelA is either 34 or 43 amino acids and 27 or 46 amino acids, respectively. The sequences encoding these sps were fused to the xylanase A gene (xlnA). Clones harboring the short sps of ManA and CelA produced as much xylanase as the clone with the control wild-type sp sequence of XlnA. In clones containing the long sps of ManA and CelA, the XlnA production was enhanced 1.5- and 2.5-fold, respectively. These XlnA yields are reduced by half and one third respectively when the internal initiation codons of the long sp sequences of ManA and CelA are mutated. Since these clones exhibited the same transcription levels, the results indicate that both RBSs are used concomitantly in S. lividans to increase the enzyme production at the translational level. However, when the short and long sps of ManA were fused to the long CelA sp sequence, giving constructs containing respectively 3 and 4 RBSs, a decrease in xylanase production was observed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Gilbert, M., Morosoli, R., Shareck, F., and Kluepfel, D. 1995. Production and secretion of proteins by Streptomycetes. Crit. Rev. Biotechnol. 15: 13–39.
Shareck, F., Roy, C., Yaguchi, M., Morosoli, R., and Kluepfel, D. 1991. Sequences of three genes specifying xylanases in Streptomyces lividans . Gene 107: 75–83.
Bertrand, J.L., Morosoli, R., Shareck, F., and Kluepfel, D. 1989. Expression of the xylanase gene of Streptomyces lividans and production of the enzyme on natural substrates. Biotechnol. Bioeng. 33: 791–794.
Pugsley, A.P. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57: 50–108.
von Heijne, G. and Abrahamsen, L. 1989. Species-specific variation in signal peptide design. Implications for protein secretion in foreign host. FEBS Lett. 244: 439–446.
Park, J.S., Horinouchi, S., and Beppu, T. 1991. Characterization of leader peptide of an endo-type cellulase produced by alkalophilic Streptomyces strain . Agric. Biol. Chem. 55: 1745–1750.
Pag´, N., Kluepfel, D., Shareck, F., and Morosoli, R. 1996. Effect of signal peptide alterations and replacement on export of xylanase A in Streptomyces lividans . Appl. Environ. Microbiol. 62: 109–114.
Arcand, N., Kluepfel, D., Paradis, F.W., Morosoli, R., and Shareck, F. 1993. β-mannanase of Streptomyces lividans 66: cloning and DNA sequence of the manA gene and characterization of the enzyme. Biochem. J. 290: 857–863.
Théberge, M., Lacaze, P., Shareck, F., Morosoli, R., and Kluepfel, D. 1992. Purification and characterization of an endoglucanase from Streptomyces lividans 66 and DNA sequence of the gene. Appl. Environ. Microbiol. 58: 815–820.
Masson, J.Y., Boucher, I., Neugebauer, N.A., Ramotar, D., and Brzezinski, R. 1995. A new chitosanase from a Nocardioides sp. is a third member of glycosyl hydrolase family 46. Microbiology 141: 2629–2635.
Taguchi, S., Nishiyama, K., Kumagai, I., Momose, H., and Miura, K. 1991. Relationship between utilization of dual translational initiation signals and protein processing in Streptomyces . Mol. Gen. Genet. 226: 328–331.
Rayner, G., Willard, J.M.A., and Schottel, J.L. 1990. Cloning, sequencing and regulation of expression of an extracellular esterase gene from plant pathogen Streptomyces scabies . J. Bacteriol. 172: 7020–7026.
Strohl, W.R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20: 961–974.
Gold, L. 1988. Posttranscriptional regulatory mechanism in Escherichia coil . Annu. Biochem. 57: 199–233.
Hurtubise, Y., Shareck, F., Kluepfel, D., and Morosoli, R. 1995. A cellulase/ xylanase negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17: 367–377.
Hopwood, D.A., Bibb, M.J., Chater, K.F., Kieser, T., Bruton, C.J., Kieser, H.M., Lydiate, D.J., Smith, C.P., Ward, J.M., and Schrempf, H. 1985. Genetic Manipulation of Streptomyces: A Laboratory Manual. The John Innes Foundation, Norwich, United Kingdom.
Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Derewenda, U., Swenson, L., Green, R., Wei, Y., Morosoli, R., Shareck, F., Kluepfel, D., and Derewenda, Z.S. 1994. Crystal structure, at 2.6-Å resolution of the Streptomyces lividans xylanase A, a member of the F family of B-1,4-D-glycanases. J. Biol. Chem. 269: 20811–20814.
Kunkel, T. 1985. Rapid and efficient site-specific mutagenesis without pheno-typic selection. Proc. Natl. Acad. Sci. USA 82: 488–492.
Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74: 5463–5467.
Kluepfel, D., Daigneault, N., Morosoli, R., and Shareck, F. 1992. Purification and characterization of a new xylanase (xylanase C) produced by Streptomyces lividans 66. Appl. Microbiol. Biotechnol. 36: 626–631.
Lowry, O.H., Rosebrough, N.J., Farr, A.L., and Randall, R.J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275.
Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685.
Towbin, H., Staehelin, T., and Gordon, J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Pagé, N., Kluepfel, D., Shareck, F. et al. Increased xylanase yield in Streptomyces lividans: Dependence on number of ribosome-binding sites. Nat Biotechnol 14, 756–759 (1996). https://doi.org/10.1038/nbt0696-756
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0696-756
This article is cited by
-
Droplet-based microfluidic platform for high-throughput screening of Streptomyces
Communications Biology (2021)
-
Actinomycetes biosynthetic potential: how to bridge in silico and in vivo?
Journal of Industrial Microbiology and Biotechnology (2014)
-
p-Hydroxycinnamic acid production directly from cellulose using endoglucanase- and tyrosine ammonia lyase-expressing Streptomyces lividans
Microbial Cell Factories (2013)
-
Optimized expression of an acid xylanase from Aspergillus usamii in Pichia pastoris and its biochemical characterization
World Journal of Microbiology and Biotechnology (2008)
-
Characterization, gene cloning and expression of new xylanase XYNB with high specific activity
Chinese Science Bulletin (2003)