Abstract
Interventions that delay ageing mobilize mechanisms that protect and repair cellular components1,2,3, but it is unknown how these interventions might slow the functional decline of extracellular matrices4,5, which are also damaged during ageing6,7. Reduced insulin/IGF-1 signalling (rIIS) extends lifespan across the evolutionary spectrum, and in juvenile Caenorhabditis elegans also allows the transcription factor DAF-16/FOXO to induce development into dauer, a diapause that withstands harsh conditions1,2. It has been suggested that rIIS delays C. elegans ageing through activation of dauer-related processes during adulthood2,8,9, but some rIIS conditions confer robust lifespan extension unaccompanied by any dauer-like traits1,10,11. Here we show that rIIS can promote C. elegans longevity through a program that is genetically distinct from the dauer pathway, and requires the Nrf (NF-E2-related factor) orthologue SKN-1 acting in parallel to DAF-16. SKN-1 is inhibited by IIS and has been broadly implicated in longevity12,13,14, but is rendered dispensable for rIIS lifespan extension by even mild activity of dauer-related processes. When IIS is decreased under conditions that do not induce dauer traits, SKN-1 most prominently increases expression of collagens and other extracellular matrix genes. Diverse genetic, nutritional, and pharmacological pro-longevity interventions delay an age-related decline in collagen expression. These collagens mediate adulthood extracellular matrix remodelling, and are needed for ageing to be delayed by interventions that do not involve dauer traits. By genetically delineating a dauer-independent rIIS ageing pathway, our results show that IIS controls a broad set of protective mechanisms during C. elegans adulthood, and may facilitate elucidation of processes of general importance for longevity. The importance of collagen production in diverse anti-ageing interventions implies that extracellular matrix remodelling is a generally essential signature of longevity assurance, and that agents promoting extracellular matrix youthfulness may have systemic benefit.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kenyon, C. J. The genetics of ageing. Nature 464, 504–512 (2010)
Shore, D. E. & Ruvkun, G. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 23, 409–420 (2013)
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013)
Flurkey, K., Papaconstantinou, J., Miller, R. A. & Harrison, D. E. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc. Natl Acad. Sci. USA 98, 6736–6741 (2001)
Wilkinson, J. E. et al. Rapamycin slows aging in mice. Aging Cell 11, 675–682 (2012)
Myllyharju, J. & Kivirikko, K. I. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 20, 33–43 (2004)
Toyama, B. H. & Hetzer, M. W. Protein homeostasis: live long, won't prosper. Nature Rev. Mol. Cell Biol. 14, 55–61 (2013)
Partridge, L. & Harvey, P. H. Gerontology. Methuselah among nematodes. Nature 366, 404–405 (1993)
McElwee, J. J., Schuster, E., Blanc, E., Thomas, J. H. & Gems, D. Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279, 44533–44543 (2004)
Gems, D. et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150, 129–155 (1998)
Arantes-Oliveira, N., Berman, J. R. & Kenyon, C. Healthy animals with extreme longevity. Science 302, 611 (2003)
Sykiotis, G. P. & Bohmann, D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci. Signal 3, re3 (2010)
Tullet, J. M. et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038 (2008)
Robida-Stubbs, S. et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15, 713–724 (2012)
Libina, N., Berman, J. R. & Kenyon, C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489–502 (2003)
Wolkow, C. A., Kimura, K. D., Lee, M. S. & Ruvkun, G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290, 147–150 (2000)
Narasimhan, S. D. et al. PDP-1 links the TGF-β and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet. 7, e1001377 (2011)
Oliveira, R. P., Porter Abate, J. & Dilks, K. et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8, 524–541 (2009)
Pang, S., Lynn, D. A., Lo, J. Y., Paek, J. & Curran, S. P. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nature Commun. 5, 5048 (2014)
Herndon, L. A. et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419, 808–814 (2002)
Varani, J. et al. Decreased collagen production in chronologically aged skin: roles of age-dependent alteration in fibroblast function and defective mechanical stimulation. Am. J. Pathol. 168, 1861–1868 (2006)
Budovskaya, Y. V. et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134, 291–303 (2008)
Argmann, C. et al. Pparγ2 is a key driver of longevity in the mouse. PLoS Genet. 5, e1000752 (2009)
Johnson, S. C., Rabinovitch, P. S. & Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345 (2013)
Vilchez, D. et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268 (2012)
Seeger-Nukpezah, T. & Golemis, E. A. The extracellular matrix and ciliary signaling. Curr. Opin. Cell Biol. 24, 652–661 (2012)
Munger, J. S. & Sheppard, D. Cross talk among TGF-β signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb. Perspect. Biol. 3, a005017 (2011)
Fu, H. L. et al. Discoidin domain receptors: unique receptor tyrosine kinases in collagen-mediated signaling. J. Biol. Chem. 288, 7430–7437 (2013)
Baumann, L. Skin ageing and its treatment. J. Pathol. 211, 241–251 (2007)
Tian, X. et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349 (2013)
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974)
Kim, T. H. et al. Tyrosylprotein sulfotransferase regulates collagen secretion in Caenorhabditis elegans. Mol. Cells 29, 413–418 (2010)
McKay, S. J. et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 68, 159–169 (2003)
Link, C. D. & Johnson, C. J. Reporter transgenes for study of oxidant stress in Caenorhabditis elegans. Methods Enzymol. 353, 497–505 (2002)
Hong, L. et al. MUP-4 is a novel transmembrane protein with functions in epithelial cell adhesion in Caenorhabditis elegans. J. Cell Biol. 154, 403–414 (2001)
Kwon, E. S., Narasimhan, S. D., Yen, K. & Tissenbaum, H. A. A new DAF-16 isoform regulates longevity. Nature 466, 498–502 (2010)
Pujol, N. et al. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr. Biol. 18, 481–489 (2008)
An, J. H. & Blackwell, T. K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17, 1882–1893 (2003)
Suzuki, Y., Morris, G. A., Han, M. & Wood, W. B. A cuticle collagen encoded by the lon-3 gene may be a target of TGF-β signaling in determining Caenorhabditis elegans body shape. Genetics 162, 1631–1639 (2002)
Calfon, M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002)
Benedetti, C., Haynes, C. M., Yang, Y., Harding, H. P. & Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174, 229–239 (2006)
Hao, L. et al. The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev. Dyn. 235, 1469–1481 (2006)
Henderson, S. T. & Johnson, T. E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11, 1975–1980 (2001)
Thein, M. C. et al. Caenorhabditis elegans exoskeleton collagen COL-19: an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev. Dyn. 226, 523–539 (2003)
Arantes-Oliveira, N., Apfeld, J., Dillin, A. & Kenyon, C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505 (2002)
Neal, S. J., Kim, K. & Sengupta, P. Quantitative assessment of pheromone-induced dauer formation in Caenorhabditis elegans. Methods Mol. Biol. 1068, 273–283 (2013)
Curran, S. P. & Ruvkun, G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3, e56 (2007)
Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. & Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, research0002 (2001)
Rual, J. F. et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 (2004)
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001)
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998)
Saldanha, A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004)
Dennis, G., Jr et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, 3 (2003)
Pirooznia, M., Nagarajan, V. & Deng, Y. GeneVenn - a web application for comparing gene lists using Venn diagrams. Bioinformation 1, 420–422 (2007)
Pavesi, G., Mereghetti, P., Mauri, G. & Pesole, G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 32, W199–W203 (2004)
Elemento, O., Slonim, N. & Tavazoie, S. A universal framework for regulatory element discovery across all genomes and data types. Mol. Cell 28, 337–350 (2007)
Thomas-Chollier, M. et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 36, W119–W127 (2008)
Glover-Cutter, K., Kim, S., Espinosa, J. & Bentley, D. L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nature Struct. Mol. Biol. 15, 71–78 (2008)
Hoogewijs, D., Houthoofd, K., Matthijssens, F., Vandesompele, J. & Vanfleteren, J. R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9, 9 (2008)
Huang, C., Xiong, C. & Kornfeld, K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 101, 8084–8089 (2004)
Zito, E., Hansen, H. G., Yeo, G. S., Fujii, J. & Ron, D. Endoplasmic reticulum thiol oxidase deficiency leads to ascorbic acid depletion and noncanonical scurvy in mice. Mol. Cell 48, 39–51 (2012)
Kage-Nakadai, E. et al. Two very long chain fatty acid acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS ONE 5, e8857 (2010)
Hertweck, M., Gobel, C. & Baumeister, R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell 6, 577–588 (2004)
Patel, D. S. et al. Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics 178, 931–946 (2008)
Dillin, A., Crawford, D. K. & Kenyon, C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830–834 (2002)
Apfeld, J. & Kenyon, C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95, 199–210 (1998)
Timmons, L., Tabara, H., Mello, C. C. & Fire, A. Z. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol. Biol. Cell 14, 2972–2983 (2003)
Kennedy, S., Wang, D. & Ruvkun, G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645–649 (2004)
Blackwell, T. K., Bowerman, B., Priess, J. R. & Weintraub, H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science 266, 621–628 (1994)
Niu, W. et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 21, 245–254 (2011)
Simmer, F. et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317–1319 (2002)
Jones, S. J. et al. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 11, 1346–1352 (2001)
Hillier, L. W. et al. Genomics in C. elegans: so many genes, such a little worm. Genome Res. 15, 1651–1660 (2005)
Shaw, W. M., Luo, S., Landis, J., Ashraf, J. & Murphy, C. T. The C. elegans TGF-β dauer pathway regulates longevity via insulin signaling. Curr. Biol. 17, 1635–1645 (2007)
Hedgecock, E. M. & Herman, R. K. The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics 141, 989–1006 (1995)
Schultz, R. D. & Gumienny, T. L. Visualization of Caenorhabditis elegans cuticular structures using the lipophilic vital dye DiI. J. Vis. Exp. 59, e3362 (2012)
Kim, T. H., Kim, Y. J., Cho, J. W. & Shim, J. A novel zinc-carboxypeptidase SURO-1 regulates cuticle formation and body morphogenesis in Caenorhabditis elegans. FEBS Lett. 585, 121–127 (2011)
Viswanathan, M., Kim, S. K., Berdichevsky, A. & Guarente, L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell 9, 605–615 (2005).
Menzel, R. et al. The nematode Caenorhabditis elegans, stress and aging: identifying the complex interplay of genetic pathways following the treatment with humic substances. Front. Genet. 3, 50 (2012)
Pietsch, K. et al. Meta-analysis of global transcriptomics suggests that conserved genetic pathways are responsible for quercetin and tannic acid mediated longevity in C. elegans. Front. Genet. 3, 48 (2012)
Gusarov, I. et al. Bacterial nitric oxide extends the lifespan of C. elegans. Cell 152, 818–830 (2013)
Schmeisser, S. et al. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Mol. Metabol. 2, 1–11 (2013)
Staab, T. A. et al. The conserved SKN-1/Nrf2 stress response pathway regulates synaptic function in Caenorhabditis elegans. PLoS Genet. 9, e1003354 (2013)
Iser, W. B., Wilson, M. A., Wood, W. H., III, Becker, K. & Wolkow, C. A. Co-regulation of the DAF-16 target gene, cyp-35B1/dod-13, by HSF-1 in C. elegans dauer larvae and daf-2 insulin pathway mutants. PLoS ONE 6, e17369 (2011)
Zarse, K. et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 15, 451–465 (2012)
Halaschek-Wiener, J. et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 15, 603–615 (2005)
Mair, W. et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature 470, 404–408 (2011)
Greer, E. L. et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466, 383–387 (2010)
Cristina, D., Cary, M., Lunceford, A., Clarke, C. & Kenyon, C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 5, e1000450 (2009)
Chen, D. et al. Germline signaling mediates the synergistically prolonged longevity produced by double mutations in daf-2 and rsks-1 in C. elegans. Cell Rep. 5, 1600–1610 (2013)
Chen, S. et al. The conserved NAD(H)-dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc. Natl Acad. Sci. USA 106, 1496–1501 (2009)
Huang da. W, Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009)
Huang, da. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Acknowledgements
We thank C. Kenyon, S. Mitani, and J. Shim for strains, P. Sengupta for dauer pheromone, C. Obieglo, L. Moronetti, M. Bland, and K. Patel for assistance, and J. Apfeld, E. Greer, C. Kenyon, W. Mair, and Blackwell laboratory members for discussions or comments on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). The work was supported by funding from the National Institutes of Health to T.K.B. (GM062891), C.T.M. (New Innovator), and J.P.A. (5T32DK007260), a Diabetes Research Center award to the Joslin Diabetes Center (P30DK036836), and fellowships from the National Science Foundation to J.N.L., and the Swiss National Science Foundation (PBSKP3_140135) to C.Y.E.
Author information
Authors and Affiliations
Contributions
All authors participated in designing the experiments, and analysing and interpreting the data. J.N.L. and J.P.A. obtained samples for microarray analysis, performed the microarray experiments, analysed the expression profiling data, and performed the lifespan studies in Extended Data Fig. 2f–h and Supplementary Table 4. C.Y.E. performed all other experiments. C.Y.E. and T.K.B. wrote the manuscript in consultation with the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Analyses of rIIS under dauer-independent and dauer-predisposed conditions.
a, Data from this study illustrating that rIIS longevity dependence upon skn-1 correlates with low dauer pathway activity, not temperature or percentage increase in mean lifespan extension (*described in the Supplementary Discussion). b, Partial schematic of the IIS pathway in C. elegans. Insulin-like peptides (ins) bind to DAF-2, leading to activation of the AKT-1/2 and possibly SGK-1 kinases1,13,63, which phosphorylate DAF-16 and SKN-1. Class 1 daf-2 mutations are typically located on the extracellular portion of DAF-2, whereas most class 2 mutations affect its intracellular domains64. c, Mutant phenotypes of daf-2. Red indicates penetrance specifically at higher temperatures (Supplementary Discussion). d, The class 2 (dauer-related) daf-2 trait of reduced body length is skn-1-independent. Each dot represents an animal, with P values determined by one-way analysis of variance (ANOVA) with post hoc Tukey’s test. e, Dependence of dauer-independent daf-2 longevity on adulthood skn-1. daf-2(e1370) lifespan extension requires skn-1 when the temperature is downshifted to 15 °C specifically during adulthood (blue). For additional information see Supplementary Table 2. f, The skn-1 dependence of daf-2(e1370) longevity at 20 °C when DAF-16 is expressed specifically in the intestine (strain description in Extended Data Table 1). g, Intestine-specific DAF-16 expression fails to rescue a class 2 dauer-like trait (immobility) in daf-2(e1370). h–j, Condition-specific induction of dauer by daf-2 RNAi. The daf-2 RNAi fails to induce dauer entry even at 25 °C (j), although some dauers are seen under more extreme conditions (27 °C)65. The activity of IIS and DAF-16 in neurons is critical for dauer regulation15,16,66, and in the wild-type RNAi is comparatively ineffective in neurons67, suggesting that the extremely weak dauer propensity of daf-2 RNAi might derive from a failure to reduce IIS sufficiently in neurons. Supporting this idea, daf-2 RNAi induced dauer entry even at 20 °C in eri-1(mg366); lin-15B(n744) mutants, in which neuronal RNAi is robust68 (h). N > 100 for each condition, two merged trials. k–n, Robust SKN-1 and DAF-16 nuclear localization under conditions of dauer inactivity. SKN-1 nuclear accumulation is inhibited comparably by IIS at 15 and 20 °C. SKN-1 is constitutively localized to ASI neuron nuclei in wild-type animals, and accumulates in intestinal nuclei in daf-2(e1370)13. k, Extent of IIS reduction from daf-2(e1370) at 15 °C, indicated by nuclear SKN-1::GFP. Chevrons indicate intestinal nuclei; scale bar, 20 μm. SKN-1::GFP (LD001) in intestinal nuclei is quantified in l, m. N > 60 for each condition and trial, three merged trials with P values determined by χ2 test. Nuclear accumulation was scored as in Methods. n, The daf-2 RNAi comparably induces SKN-1::GFP (LD001) and DAF-16::GFP (TJ356) intestinal nuclear localization at 15 and 20 °C. N > 60 for each condition, one trial with all experimental conditions done in parallel. o, Comparable nuclear accumulation of DAF-16f::GFP (lpIs14) induced by daf-2 RNAi and daf-2(e1370) at 15 and 20 °C. N > 60 for each condition, one trial performed in parallel. p, q, Induction of dauer development (p) and dauer-like traits (skn-1-independent) (q) by the crude dauer pheromone preparation used in lifespan assays (Fig. 1e, Extended Data Table 1 and Supplementary Table 2). In p, N > 100 for each condition, one trial. In q, N = 30 for each condition, three merged trials. For h–j, l–o, P values were determined by χ2 test; n.s., not significant, *P < 0.05, **P < 0.001, ***P < 0.0001.
Extended Data Figure 2 Identification of SKN-1-regulated daf-2(−) genes.
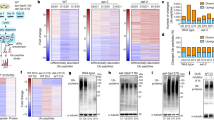
a, Heatmap of 429 genes identified by SAM as significantly upregulated by SKN-1 in daf-2 mutants. b, Four hundred and seventy-seven genes identified by SAM as significantly downregulated by SKN-1 in daf-2 mutants (Supplementary Table 3). The SKN-1-downregulated daf-2(−) set was enriched for genes involved in ubiquitin-mediated proteolysis (E3 ligase/SCF, F-box; Supplementary Table 8). Columns represent biological samples. Blue, down; black, unregulated. c, Confirmation of microarray data for SKN-1-upregulated daf-2(−) genes by qPCR at 15 °C. One and three biological replicates were analysed in the left and right panels, respectively. SAM scores are in Supplementary Table 3. Data are mean ± s.e.m. *P < 0.05, **P < 0.001, ***P < 0.0001 relative to daf-2, determined by one sample t-test, two-tailed, hypothetical mean of 1. d, e, Enrichment of SKN-1 binding sites upstream of SKN-1-regulated daf-2(−) genes. An unbiased search using the Weeder and FIRE algorithms did not detect any overrepresented form of the consensus SKN-1 binding motif (WWTRTCAT) (W = A/T, R = G/A)69. Given the degeneracy of this motif, we used RSATools to perform a directed search of 600 bp upstream of SKN-1 upregulated (d) and downregulated (e) genes. This search window was based upon the location of SKN-1 binding sites identified by genome-wide chromatin immunoprecipitation followed by sequencing (ChIP-seq) using transgenically expressed SKN-1 (ref. 70). A SKN-1 motif was detected at only 13% of a random sample of 10,000 genes, but at 37% and 24% of the SKN-1-upregulated (out of 429 genes) and downregulated genes (out of 477 genes), respectively. f, Importance of SKN-1-upregulated daf-2(−) genes for daf-2(e1368) lifespan. The class 1 daf-2 allele e1368 is partly dependent upon skn-1 for lifespan extension at 20 °C (Extended Data Table 1)13. Adult RNAi against 5 of 12 genes tested reduced daf-2(e1368) lifespan at 20 °C. g, h, Several SKN-1-downregulated daf-2(−) genes decrease lifespan. Knockdown was performed in the RNAi-sensitive strain rrf-3(pk1426)71. g, Genes for which RNAi knockdown increased lifespan, from 12 that were analysed without regard to their function. h, Analysis of six Skp1-related genes, an overrepresented category among SKN-1-downregulated daf-2(−) genes (Supplementary Table 8). Only genes that affected lifespan are shown. Other data and all statistics are in Supplementary Table 6. For 15 other SKN-1-downregulated daf-2(−) genes, it has been shown previously that RNAi increases lifespan (Supplementary Table 5). Parts f and g each show a single trial, and a composite of three trials is shown in h. In g the negative RNAi control is elpc-4(RNAi) instead of L4440. Mean lifespan (in days) is indicated for each gene. i, Overlap between the daf-2(−); SKN-1-dependent upregulated gene set (429 genes, this study) and a set of genes preferentially upregulated in dauers (358 genes)72. The overlap of six genes was not significant (P = 0.6391 by two-sided χ2 test). The number of genes that were present in neither set (no/no) was determined by subtracting the total number in both gene sets from the total number of genes encoded in C. elegans 19,735 (ref. 73). j, Overlaps between SKN-1-regulated daf-2(−) and DAF-16-regulated daf-2(−) gene sets74. For both up- and downregulated daf-2(−) genes, overlaps between the SKN-1- and DAF-16-regulated sets were significant (P < 0.0001 determined by two-sided χ2 test). Moreover, hierarchical clustering identified additional SKN-1-upregulated daf-2(−) genes that were also upregulated by DAF-16 even though they were not present in this list of highest-confidence DAF-16-regulated genes (l). The number of genes that were in neither set (no/no) was determined as in i. k, Hierarchical clustering of SKN-1-downregulated daf-2(−) gene sets with DAF-16-regulated genes. SKN-1-regulated genes identified here were queried as to how they were influenced by DAF-16 in a comparison of daf-2(e1370) versus daf-16(mu86); daf-2(e1370) animals raised at 20 °C (ref. 74). Three hundred and ninety-three SKN-1-upregulated daf-2(−) genes that were present in this DAF-16-regulated data set are shown. Most SKN-1-downregulated daf-2(−) genes did not appear to be regulated by DAF-16. l, Hierarchical clustering of SKN-1-upregulated daf-2(−) genes with DAF-16-regulated genes that were identified by comparing daf-2(e1370) versus daf-16(mu86); daf-2(e1370) at 20 °C (ref. 74). Two hundred and seventy-two SKN-1-upregulated daf-2(−) genes that were present in this DAF-16-regulated data set are shown, 46% of which were upregulated by both SKN-1 and DAF-16. Yellow, up; blue, down; black, unregulated. m–t, Effects of SKN-1 and DAF-16 on individual genes in response to daf-2 RNAi at 20 °C. A qPCR analysis of skn-1(zu67), daf-16(mgDf47), and daf-16(mgDf47); skn-1(zu67) double mutants indicated that many genes are upregulated by daf-2(RNAi) (red) in a skn-1-dependent manner, but also that these genes vary in how they are affected by DAF-16. DAF-16 and SKN-1 increased activity of gst-4, col-65, and col-176, but DAF-16 seemed to downregulate dod-24, nas-7, and F55G11.2. All of these genes except ins-7 were identified in our daf-2; skn-1 data sets. For each condition, three biological samples of 200 worms each were analysed by qPCR. All data are mean ± s.e.m. *P < 0.05, **P < 0.001, ***P < 0.0001 relative to wild-type RNAi control, determined by one sample t-test, two-tailed, hypothetical mean of 1.
Extended Data Figure 3 Analyses of SKN-1-regulated daf-2(−) genes.
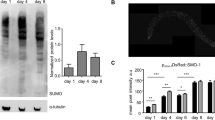
a–f, SKN-1-upregulated (a–c) and downregulated (d–f) daf-2(−) gene sets were examined by hierarchical clustering to determine how they were previously found to be affected by SKN-1 under unstressed or oxidative stress conditions18. t-BOOH, tert-butyl hydroperoxide. g, Proportional Venn diagrams show comparisons of SKN-1-upregulated genes identified under daf-2(−), normal, or arsenite treatment conditions18 (Supplementary Table 7). In each case, L4 larvae were analysed to avoid embryogenesis effects. Heatmaps are shown in a–f. h, The SKN-1-upregulated daf-2(−) collagen col-89 is expressed in neurons and the intestine, but not in hypodermis. Transgenic Pcol-89::GFP (BC12533) at day 8 of adulthood is shown. Anterior to the left, ventral side down; scale bar, 100 μm. i–k, SKN-1-mediated collagen gene activation in day 8 daf-2(RNAi) adults. Adulthood daf-2 knockdown (i) activated a Pcol-12::dsRED reporter (j; scale bar, 100 μm). k, The skn-1 dependence of Pcol-12::dsRED expression. EV, empty RNAi vector. N > 60 for each condition, three merged trials, with P value by χ2 test (*P < 0.05; ***P < 0.0001; n.s., not significant).
Extended Data Figure 4 rIIS delays age-associated decline in collagen expression.
a, Age-associated decline in expression of selected collagen and SKN-1-dependent detoxification genes. Eighty-eight collagens are among many genes that decline in expression as C. elegans ages22. Fifty of these age-downregulated genes were in our SKN-1 upregulated daf-2(−) gene set, including 27 collagen genes (Supplementary Table 10). These daf-2(−); SKN-1-dependent collagens were neither flanked by SKN-1 binding sites nor bound by SKN-1 in a genome-wide survey (Supplementary Table 9)70, suggesting that they are regulated by SKN-1 indirectly. The average Cy5-labelled cDNA values of day 2–11 adults (indicated as ‘exp’) are plotted in binary logarithm (log2) relative to cy3-labelled reference cDNA from mixed stage hermaphrodites (indicated as ‘ref’). Data are from ref. 22. The nit-1, gst-4, and F56D5.3 genes are predicted to encode a nitrilase, glutathione S-transferase, and NADPH oxidoreductase, respectively (WormBase). b, Expression of SKN-1-regulated collagen and oxidative stress response genes (nit-1 and gst-4) are maintained during ageing in daf-2(RNAi) animals. One-day-old adult wild-type (N2) animals were placed on either empty vector control (L4440) (black) or daf-2 RNAi (red) at 20 °C. mRNA was harvested at days 3, 6, and 8. mRNA levels are shown relative to wild-type (N2) day 3 adults on empty vector control (L4440) RNAi and are represented as mean ± s.e.m. For each condition, two biological samples of more than 100 worms each were analysed by qPCR. For each gene, the statistical difference of relative mRNA expression levels between L4440 and daf-2(RNAi) treatment over the time course (days 3, 6, 8) is shown by two-way ANOVA (repeated measures). c, Adulthood knockdown of SKN-1-upregulated collagens did not affect wild-type lifespan For statistics and additional trials, see Extended Data Table 3 and Supplementary Table 13. d, Importance of SKN-1-upregulated collagens for daf-2(RNAi) longevity. Adulthood RNAi knockdown of daf-2 combined with collagens or skn-1 at 20 °C is shown. GFP was the RNAi control. For statistics and additional data, see Supplementary Table 13. e, Suppression of daf-2(e1370) but not wild-type longevity at 15 °C by the collagen mutation dpy-1(e1), which affects the cuticle31,75, but was not present in our SKN-1-regulated gene set. For details and statistics see Supplementary Table 13. f, Longevity of daf-2(e1370) at 15 °C requires the SKN-1-upregulated extracellular proteases asp-14 and suro-1, along with cuticle integrity genes acs-20 and acs-22 (ref. 62), suggesting a general importance of ECM gene expression. For details and statistics see Supplementary Table 13.
Extended Data Figure 5 Adulthood knockdown of collagens important for longevity does not affect morphology of cuticle-associated structures.
a, Schematic cross-section of C. elegans illustrating the proximity of the cuticle (black), hypodermis (red), basal lamina (blue), and body-wall muscles (purple). Annuli, furrow, and alae are characteristic cuticle structures. b–j, Adulthood RNAi against SKN-1-upregulated daf-2(−) collagens does not affect cuticle morphology. b–f, One-day-old wild-type animals were exposed to either empty vector (control) or the indicated RNAi clone by feeding. Ten days later, animals were incubated in DiI for 16 h; the cuticle was imaged as described in ref. 76. N > 30 animals per condition scored, with typical images shown. Scale bar, 10 μm. g–j, Cuticle morphology revealed by the collagen COL-19, detected by a translational fusion protein (kaIs12 [COL-19::GFP]). We did not identify col-19 as being regulated by daf-2 and skn-1, and daf-2(RNAi) did not detectably alter COL-19::GFP levels (not shown). k–n, Adulthood knockdown of SKN-1-upregulated daf-2(−) collagens does not affect the pattern of chEx1682 QUA-1::GFP, a marker of cuticle adhesion. QUA-1 encodes a hedgehog-related protein required for moulting, cuticle adhesion, and alae formation42. o–r, Adulthood RNAi against SKN-1-upregulated daf-2(−) collagens does not affect the pattern of muscle–hypodermis–cuticle adhesion, as indicated by upIs1 MUP-4::GFP. MUP-4 is a transmembrane protein that is part of a complex that attaches hypodermis and muscles to the cuticle35. s–v, Adulthood collagen knockdown does not affect mitochondrial morphology in muscle. For g–v, animals were placed on RNAi at the first day of adulthood and scored and imaged at day 8 of adulthood. N > 30 animals per condition scored, with typical images shown. Scale bar, 10 μm.
Extended Data Figure 6 Phenotypic analyses of collagens important for longevity.
a, Adulthood col-120 knockdown does not affect daf-2(e1370) body size at 15 °C. The daf-2(e1370) animals were placed on RNAi food as day 1 adults, and at day 10 body size, pharyngeal pumping, and lipofuscin levels were scored in parallel in the same animals (N > 30; one trial; see Fig. 4a, b). b, c, Adulthood knockdown of SKN-1-upregulated collagens does not alter barrier function. b, Upper panel: animals were placed on RNAi food on adulthood day 1, and at day 9 were incubated in 1 μg ml−1 Hoechst 33342, which is membrane-permeable but cuticle-impermeable. For details see Methods, adapted from ref. 62. Lower panel: barrier permeability was not affected by daf-2 mutation or collagen knockdown. Permeability was assessed by nuclear Hoechst staining in the tail62 (N > 50 per condition; one trial). Approximately half of the animals in each group showed nuclear staining in the tail that is likely to have arisen through uptake in the intestine, as suggested by the high levels of intestinal Hoechst staining (c). Uptake through the cuticle would have resulted in a much wider distribution of stained nuclei. c, Representative pictures of quantification categories. Arrow indicates Hoechst-stained tail nuclei. Scale bar, 50 μm. d, Adulthood knockdown of col-120 did not sensitize to hypertonic stress. Day 1 adult wild-type animals were placed RNAi food for 3 days, then on plates containing food and high concentrations of salt for 24 h before assay (NaCl: 450 mM, 500 mM, 600 mM; N > 60 per condition; two trials). e, Adulthood knockdown of SKN-1-upregulated collagens did not impair body movement. Neither the frequency nor morphology (not shown) of body movement was affected. In parallel, the daf-2 RNAi control increased movement frequency because these animals were chronologically younger. (**P < 0.001, one-way ANOVA post hoc Tukey’s test compared with empty RNAi vector.) f, Adulthood collagen RNAi did not increase vulval rupturing during ageing. The bar graph shows the mean ± s.e.m. percentage of exploded worms that were censored during lifespan assays (Extended Data Table 3 and Supplementary Table 13). g–i, Adulthood col-120 knockdown did not induce unfolded protein, heat-shock stress, or oxidative stress responses. In g, daf-2(e1370) mutants were placed on RNAi food as day 1 adults, and assayed at day 8 (upper panel). Relative levels of these stress response gene mRNAs were determined by qPCR (two independent trials, each with 200 worms per condition). h, Adulthood collagen RNAi does not activate the oxidative stress response marker Pgst-4::GFP34, assayed after 4 and 8 days of RNAi. As a control, daf-2 RNAi induced SKN-1 to increase gst-4 expression (Fig. 2a). i, Adulthood collagen RNAi does not activate the unfolded protein response marker Phsp-4::GFP40. j–l, Importance of collagens for oxidative stress resistance. Day 1 adults were exposed to empty vector (EV) or RNAi food at 15 °C, then at day 3 were placed in 5 mM arsenite (As) and scored for survival. Knockdown of collagens and other SKN-1 upregulated daf-2(−) genes sensitized to oxidative stress from arsenite; nit-1 (nitrilase), gst-4 (glutathione S-transferase), F56D5.3 (NADPH oxidoreductase). *P < 0.05, **P < 0.01, ***P < 0.001 relative to control (empty RNAi vector), determined by one-way ANOVA with post hoc Tukey’s test. t-BOOH experiments are described in Supplementary Table 16. m, Adulthood collagen expression required for rapamycin to delay appearance of an ageing marker (pharyngeal pumping). N > 30; each dot represents an animal; ***P < 0.0001 determined with an unpaired t-test, two-tailed.
Extended Data Figure 7 Cuticle remodelling in adults
a, The collagen ROL-6 is present in the cuticle during development and early adulthood77, then largely disappears during ageing. The upper panels show the mid-body (left) and head (right) regions in an L4 animal. Day 1 adults exhibited similar levels and patterns of jgIs5 ROL-6::GFP fluorescence (not shown). Lower panels show the corresponding regions in a day 8 adult, in which jgIs5 ROL-6::GFP levels are reduced. The orange signal corresponds to gut autofluorescence. Representative images are shown; scale bar, 20 μm. N = 30 for each sample set (L4, day 1, and day 8). b, Total collagen levels are elevated in long-lived animals at the first day of adulthood. Note that these long-lived animals also maintain higher collagen levels in later life despite an age-related decline (Fig. 4d). Relative collagen levels were estimated by a hydroxyproline assay61. In daf-2 mutants, total collagen levels were elevated at both temperatures but the increase was greater at 15 °C, at which skn-1 and SKN-1-dependent collagens are required for lifespan extension (Fig. 3 and Supplementary Table 13). Temperature-sensitive glp-1(bn18) mutants were maintained at 15 °C (permissive temperature), or upshifted to 25 °C (restrictive temperature) as L1 larvae until the L4 stage, then placed at 20 °C. c, SKN-1-dependent collagen genes from the daf-2(−) set are not upregulated in 8-day-old daf-2(e1370) adults at 20 °C. Expression of these collagens remains increased at this age in daf-2(e1370) at 15 °C or after daf-2 RNAi at 20 °C (Fig. 2a and Extended Data Figs 2c and 4b), conditions in which the dauer pathway is inactive and lifespan extension is skn-1 dependent (see text). Two hundred day-8 adults were assayed in each sample, with three merged independent trials shown. d, Scoring categories for the Pcol-144::GFP reporter are shown in (e, g; Fig. 4f; scale bar, 100 μm). e, Adulthood rapamycin treatment increases col-144 promoter activity. Knockdown of col-10, col-13, or col-120 did not reduce Pcol-144::GFP levels at day 4, but significantly decreased Pcol-144::GFP levels by day 8 (g). N > 60 for each condition, two merged trials, with P value by χ2 test (#P < 0.0001 against untreated empty RNAi vector control animals). f, Dependence of the SKN-1 target gene gst-4 on adulthood SKN-1-upregulated collagen expression in daf-2(RNAi) animals. Collagen or empty vector (EV) control RNAi was initiated at day 1 of adulthood at 20 °C, together with daf-2 knockdown. g, Adulthood collagen RNAi decreases col-144 promoter activity in rapamycin-treated animals. As is seen in daf-2 mutants at 15 °C (Fig. 4f), activity of this rapamycin-activated promoter is unaffected by adulthood collagen RNAi at day 4 (e), but reduced at day 8. For f and g, N > 60 for each condition, two merged trials, with P value by χ2 test (#P < 0.0001).
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2 and 4-17, a Supplementary Discussion and additional references (see separate file for Supplementary Table 3). (PDF 731 kb)
Supplementary Table 3
The file contains a complete list of daf-2; skn-1-up- and downregulated gene sets. (XLSX 64 kb)
daf-2(e1370) (Class 2) mutants were grown and maintained at 15°C.
When maintained at 15°C, daf-2(e1370) mutants (Class 2) are mobile throughout their lifespan as shown here at day 14 of adulthood (10 min; 8x speed). (MOV 9991 kb)
daf-2(e1370) (Class 2) mutants were grown and maintained at 15°C.
When upshifted from 15°C to 20°C at the first day of adulthood, daf-2(e1370) is typical of other Class 2 mutants in that it becomes relatively immobile during much of its adult lifespan, as shown here at day 14 (10 min; 8x speed). (MOV 6661 kb)
Rights and permissions
About this article
Cite this article
Ewald, C., Landis, J., Abate, J. et al. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519, 97–101 (2015). https://doi.org/10.1038/nature14021
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14021
This article is cited by
-
ADARs regulate cuticle collagen expression and promote survival to pathogen infection
BMC Biology (2024)
-
Ovalbumin promotes innate immune response of Caenorhabditis elegans through DAF-16 and SKN-1 pathways in insulin/IGF-1 signaling
Journal of Physiology and Biochemistry (2024)
-
Unraveling effects of anti-aging drugs on C. elegans using liposomes
GeroScience (2023)
-
Aging of mesenchymal stem cell: machinery, markers, and strategies of fighting
Cellular & Molecular Biology Letters (2022)
-
Distinct and additive effects of calorie restriction and rapamycin in aging skeletal muscle
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.