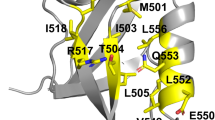
Abstract
Protein sequences evolve through random mutagenesis with selection for optimal fitness1. Cooperative folding into a stable tertiary structure is one aspect of fitness, but evolutionary selection ultimately operates on function, not on structure. In the accompanying paper2, we proposed a model for the evolutionary constraint on a small protein interaction module (the WW domain) through application of the SCA, a statistical analysis of multiple sequence alignments3,4. Construction of artificial protein sequences directed only by the SCA showed that the information extracted by this analysis is sufficient to engineer the WW fold at atomic resolution. Here, we demonstrate that these artificial WW sequences function like their natural counterparts, showing class-specific recognition of proline-containing target peptides5,6,7,8. Consistent with SCA predictions, a distributed network of residues mediates functional specificity in WW domains. The ability to recapitulate natural-like function in designed sequences shows that a relatively small quantity of sequence information is sufficient to specify the global energetics of amino acid interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Voigt, C. A., Kauffman, S. & Wang, Z. G. Rational evolutionary design: the theory of in vitro protein evolution. Adv. Protein Chem. 55, 79–160 (2000)
Socolich, M. et al. Evolutionary information for specifying a protein fold. Nature doi:10.1038/nature03991 (this issue)
Lockless, S. W. & Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 (1999)
Suel, G. M., Lockless, S. W., Wall, M. A. & Ranganathan, R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nature Struct. Biol. 10, 59–69 (2003)
Bedford, M. T., Sarbassova, D., Xu, J., Leder, P. & Yaffe, M. B. A novel pro-Arg motif recognized by WW domains. J. Biol. Chem. 275, 10359–10369 (2000)
Chen, H. I. & Sudol, M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl Acad. Sci. USA 92, 7819–7823 (1995)
Ermekova, K. S. et al. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J. Biol. Chem. 272, 32869–32877 (1997)
Lu, P. J., Zhou, X. Z., Shen, M. & Lu, K. P. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283, 1325–1328 (1999)
Zarrinpar, A. & Lim, W. A. Converging on proline: the mechanism of WW domain peptide recognition. Nature Struct. Biol. 7, 611–613 (2000)
Kanelis, V., Rotin, D. & Forman-Kay, J. D. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nature Struct. Biol. 8, 407–412 (2001)
Verdecia, M. A., Bowman, M. E., Lu, K. P., Hunter, T. & Noel, J. P. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nature Struct. Biol. 7, 639–643 (2000)
Kato, Y. et al. Common mechanism of ligand recognition by group II/III WW domains: redefining their functional classification. J. Biol. Chem. 279, 31833–31841 (2004)
Hu, H. et al. A map of WW domain family interactions. Proteomics 4, 643–655 (2004)
Otte, L. et al. WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci. 12, 491–500 (2003)
Chen, H. I. et al. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J. Biol. Chem. 272, 17070–17077 (1997)
Espanel, X. & Sudol, M. A single point mutation in a group I WW domain shifts its specificity to that of group II WW domains. J. Biol. Chem. 274, 17284–17289 (1999)
Kasanov, J., Pirozzi, G., Uveges, A. J. & Kay, B. K. Characterizing Class I WW domains defines key specificity determinants and generates mutant domains with novel specificities. Chem. Biol. 8, 231–241 (2001)
Toepert, F., Pires, J. R., Landgraf, C., Oschkinat, H. & Schneider-Mergener, J. Synthesis of an array comprising 837 variants of the hYAP WW protein domain. Angew. Chem. Int. Edn Engl. 40, 897–900 (2001)
Huang, X. et al. Structure of a WW domain containing fragment of dystrophin in complex with β-dystroglycan. Nature Struct. Biol. 7, 634–638 (2000)
Carter, P. J., Winter, G., Wilkinson, A. J. & Fersht, A. R. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell 38, 835–840 (1984)
Hidalgo, P. & MacKinnon, R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science 268, 307–310 (1995)
Dahiyat, B. I. & Mayo, S. L. De novo protein design: fully automated sequence selection. Science 278, 82–87 (1997)
Dwyer, M. A., Looger, L. L. & Hellinga, H. W. Computational design of a biologically active enzyme. Science 304, 1967–1971 (2004)
Kortemme, T. et al. Computational redesign of protein-protein interaction specificity. Nature Struct. Mol. Biol. 11, 371–379 (2004)
Kraemer-Pecore, C. M., Lecomte, J. T. & Desjarlais, J. R. A de novo redesign of the WW domain. Protein Sci. 12, 2194–2205 (2003)
Harbury, P. B., Plecs, J. J., Tidor, B., Alber, T. & Kim, P. S. High-resolution protein design with backbone freedom. Science 282, 1462–1467 (1998)
Kuhlman, B. et al. Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 (2003)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Ferguson, N., Johnson, C. M., Macias, M., Oschkinat, H. & Fersht, A. Ultrafast folding of WW domains without structured aromatic clusters in the denatured state. Proc. Natl Acad. Sci. USA 98, 13002–13007 (2001)
Delano, W. L. The PyMOL Molecular Graphics Systemhttp://www.pymol.org (2002).
Acknowledgements
We thank members of the Ranganathan laboratory for advice and critical review of the manuscript, J. P. Noel for providing the pHIS8 expression vector, and K. Voegler for contributing to this project. This study was supported by the Robert A. Welch foundation (R.R.), the Mallinckrodt Foundation Scholar Award (R.R.), NIH grants (M.B.Y.), and a Burroughs-Wellcome Career Development Award (M.B.Y.). D.M.L. was supported by a Howard Hughes Medical Institute pre-doctoral award. W.P.R. is an associate and R.R. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure Legends
Text to accompany the below Supplementary Figures. (DOC 35 kb)
Supplementary Figure S1
Binding specificity assays determined by the oriented peptide library assay for (a) 28 natural WW domains and (b) 10 artificial WW domains designed using the SCA matrix. (PDF 1571 kb)
Supplementary Figure S2
Saturation mutagenesis of the peptide ligands for the two major functional classes of WW domains identified. (PDF 6239 kb)
Supplementary Figure S3
Thermodynamic double mutant cycles in the WW domain Nedd4.3 (N39), measuring the energetic coupling between the T28A mutant and mutants at three other sites within the network of co-evolving residues (L3A, E8A, and H23A). (PDF 272 kb)
Rights and permissions
About this article
Cite this article
Russ, W., Lowery, D., Mishra, P. et al. Natural-like function in artificial WW domains. Nature 437, 579–583 (2005). https://doi.org/10.1038/nature03990
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03990
This article is cited by
-
Machine learning-aided design and screening of an emergent protein function in synthetic cells
Nature Communications (2024)
-
Nucleotide-based genetic networks: Methods and applications
Journal of Biosciences (2022)
-
Expanding functional protein sequence spaces using generative adversarial networks
Nature Machine Intelligence (2021)
-
MPL resolves genetic linkage in fitness inference from complex evolutionary histories
Nature Biotechnology (2021)
-
WW domain-binding protein 2: an adaptor protein closely linked to the development of breast cancer
Molecular Cancer (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.