Abstract
Nontypeable Haemophilus influenzae (NTHI) is a significant cause of otitis media in children and exacerbations in patients with chronic obstructive pulmonary disease. Vaccine research for NTHI has focused on the outer membrane proteins (OMPs) of NTHI. The goal of this study was to evaluate mucosal and systemic immune responses to recombinant OMP P2 (rP2) of NTHI. Enzyme-linked immunosorbent assay (ELISA) demonstrated that both mucosal and systemic routes of immunization resulted in antibodies to rP2. Whole-cell ELISA and flow cytometry indicated that mucosal immunization induced antibodies to epitopes that are on the bacterial surface of the homologous strain as well as several heterologous strains. In contrast, systemic immunization induced antibodies to non-surface exposed epitopes. These data show for the first time that mucosal immunization of mice with rP2 induces antibodies that recognize surface exposed epitopes on multiple strains, indicating that P2 is a candidate for development of a mucosal vaccine for NTHI.
Similar content being viewed by others
Introduction
Nontypeable Haemophilus influenzae (NTHI) is an important human pathogen. This nonencapsulated coccobacillus is the leading cause of lower respiratory tract infection in patients with chronic obstructive pulmonary disease.1, 2, 3 Chronic obstructive pulmonary disease is currently the fifth leading cause of death worldwide and is expected to rank fourth by 2030 (http://www.who.int/respiratory/copd/en/). NTHI is also a significant cause of otitis media in children, along with Streptococcus pneumoniae and Moraxella catarrhalis.4, 5 Recurrent ear infections have been implicated in delayed speech development and impaired cognitive ability in children,4 effects which may extend into adolescence.6 Otitis media due to NTHI has been increasing with the introduction of the seven-valent S. pneumoniae conjugate vaccine.7, 8, 9 Although antibiotics are effective against NTHI, increasing rates of antibiotic resistance are being reported.9, 10, 11 These observations underscore the need for a vaccine to prevent NTHI infection, including otitis media in children and respiratory tract infection in adults with chronic obstructive pulmonary disease.
Vaccine development for NTHI has largely focused on the outer membrane proteins (OMPs) due to their presence on the bacterial surface.12, 13 Of the proteins being evaluated for inclusion in an NTHI vaccine, OMP P2 has received little attention. OMP P2 is a homotrimeric porin constituting approximately half of the protein content of the outer membrane.14 The major drawback to the use of OMP P2 as a vaccine antigen is the sequence heterogeneity of several of the surface exposed loops among strains.15, 16, 17, 18 However, P2 also possesses characteristics to suggest that it may be an effective vaccine antigen. H. influenzae mutants deficient in P2 exhibit compromised viability,14, 19 suggesting that downregulation of expression is unlikely to occur. Furthermore, due to the abundance of P2 in the outer membrane, the protein presents multiple targets for antibody binding. Neary et al. have shown that immunization with peptides corresponding to a conserved loop of OMP P2 results in bactericidal activity against multiple strains.20, 21 These data indicate that antibodies to P2 have the potential to be protective against multiple strains. However, not all strains were recognized and the P2 amino-acid sequence alone was not sufficient to predict protection. Immunization with the full-length protein would expose the immune system to a wider range of epitopes, possibly increasing the degree of cross-reactivity of anti-P2 antibodies.
As NTHI is a mucosal pathogen, immunization strategies that emphasize both mucosal and systemic responses should be considered. Although little is known regarding the role of the mucosal immune response in preventing infection with NTHI, it has been suggested that immunoglobulin A (IgA) antibodies may aid in preventing adherence and colonization of NTHI, important preliminary events for infection.22, 23 Several studies using animal models have shown increased clearance of NTHI and other bacteria following intranasal immunization, suggesting that mucosal antibody responses are an important factor in protection.24, 25, 26, 27
The goal of this study was to test the hypothesis that immunization with OMP P2 induces antibodies that recognize epitopes on the surface of multiple strains. Purified recombinant P2 (rP2) from selected strains were generated and used to assess both mucosal and systemic immune responses by immunizing mice either intranasally (IN) or subcutaneously (SQ). Serum samples from both groups of mice were evaluated for P2-specific immunoglobulin G (IgG). In addition, saliva was collected from animals immunized IN to evaluate mucosal immune responses. Immunoassays were performed to detect antibody recognition of surface epitopes. The results show that rP2 is immunogenic when administered both mucosally and systemically. Mucosal immunization with rP2 from selected strains of NTHI induced antibodies that bind to surface exposed epitopes of multiple strains.
Results
Characterization of rP2
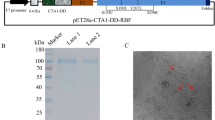
To test the hypothesis that immunization with rP2 induces cross-reactive immune responses, rP2 was generated from three strains of NTHI. Strains 13P24H1, 23P2H1, and 54P24H1 were chosen as templates based on their P2 sequences. SDS-polyacrylamide gel (PAGE) analysis of purified rP2 revealed a single band of approximately 48 kDa, the anticipated size of the P2 protein with the addition of the 6X-His tag (Figure 1). Results from circular dichroism indicated the presence of β-sheet domains, indicating that the rP2 was at least partially refolded (data not shown). A Limulus Amebocyte Lysate assay showed no detectable endotoxin.
SDS-polyacrylamide gel (PAGE) of recombinant P2. Proteins were resolved on a 12% acrylamide gel and stained with Coomassie Blue. Molecular mass markers are listed in kDa on the left.
Murine immune response to rP2
To study the immunogenicity of rP2 in mice, animals were immunized either IN with cholera toxin as adjuvant or SQ with incomplete Freund's adjuvant. Control groups included unimmunized mice and mice receiving adjuvant alone. Quantitative enzyme-linked immunosorbent assays (ELISAs) were performed to evaluate the immune response to rP2. Intranasal immunization induced P2-specific serum IgG (Table 1). In addition, anti-rP2 IgA was detected in the saliva of IN-immunized mice (Table 1). Levels of serum IgG from IN-immunized mice showed a dose-dependent effect, and this effect was most pronounced in antisera to rP213 and rP254. Levels of salivary IgA displayed greater variability than serum IgG, and a strict dose-dependent effect was not observed. These results indicate that rP2 is immunogenic in mice following mucosal immunization.
To study the effects of systemic immunization, mice were immunized by the SQ route with 20 μg of rP2 emulsified in incomplete Freund's adjuvant. Systemic immunization resulted in P2-specific serum IgG at levels approximately 1,000-fold higher than mucosal immunization (Table 2). These results indicate that rP2 is immunogenic in mice following both mucosal and systemic immunization and that systemic immunization with incomplete Freund's adjuvant induces substantially higher serum IgG levels compared with mucosal immunization with cholera toxin.
Characterization of antibodies to surface-exposed epitopes
Whole-cell ELISAs
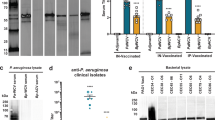
To determine if antibodies to rP2 recognize surface-exposed epitopes, whole-cell ELISA was employed. Serum from mice immunized with adjuvant alone was included in each assay as a negative control. Binding of antibodies in whole-cell ELISA was observed for antisera from IN-immunized mice, whereas no difference in binding was observed between test sera and adjuvant control sera for SQ-immunized mice (Figure 2). Background levels of antibody binding in normal mouse serum were consistently below that observed from mice immunized with adjuvant alone (data not shown). In the IN-immunized groups, the degree of binding in whole-cell ELISA varied among the three immunogens, and was not always dose dependent. Antiserum to rP223 at the 50 μg dose yielded the highest level of binding to the homologous strain. In contrast, the 20 μg dose induced the highest level of antibodies to surface epitopes in mice immunized with rP213 and rP254. The observation that serum samples that have the highest levels of antibodies in whole-cell ELISA have relatively lower levels of serum IgG detected by quantitative ELISA (Table 1) indicates that these two assays recognize two distinct antibody populations. As antibodies to surface epitopes are the most likely to be protective, we chose to study antisera that exhibited the greatest degree of binding in whole-cell ELISA in our remaining experiments. These results indicate that antibodies from mice immunized mucosally recognize surface-exposed epitopes whereas those from mice immunized systemically do not.
Serum antibodies induced by SQ and IN immunization with recombinant P2 (rP2) assayed by whole-cell enzyme-linked immunosorbent assay (ELISA) using the homologous strain of nontypeable Haemophilus influenzae (NTHI). All assays were performed with a serum dilution of 1:500. Assays were performed in triplicate and results are expressed as the mean±s.e.m. P-values were calculated using Student's t-test. OD, optical density; SQ, subcutaneous; IFA, incomplete Freund's adjuvant; IN, intranasal; CT, cholera toxin.
Flow cytometry
Flow cytometry was used as a second method to evaluate binding of antibodies to the surface of NTHI. Sera from IN-immunized mice showed a significant degree of binding to surface epitopes (Figure 3), whereas sera from SQ-immunized animals showed no binding to whole bacterial cells. Results from flow cytometry correlated well with those from whole-cell ELISA, supporting the conclusion that anti-rP2 antibodies from IN-immunized mice recognize surface-exposed epitopes, whereas those from SQ-immunized mice do not.
Serum antibodies induced by SQ and IN immunization with recombinant P2 (rP2) assayed by flow cytometry with antisera raised to rP2. Flow cytometry was performed with the homologous strain in triplicate and median fluorescence intensity was determined. IN sera were tested at a 1:10 dilution. SQ sera were tested at a 1:100 dilution. Results are expressed as the mean±s.e.m. Student's t-test was used to test for significance. SQ, subcutaneous; NMS, normal mouse serum; IN, intranasal.
Specificity of the murine antibody response to rP2
Construction of P2-deficient mutants
To address the specificity of antibodies binding to surface epitopes, P2 mutants were constructed for each of the three homologous strains. Outer membrane preparations of the parent and the mutant strains showed a loss of the P2 band from the mutant, with no visible effect on the other OMPs (Figure 4a). Immunoblots using P2-specific antisera confirmed that the P2 protein is not expressed in the mutant strains (Figure 4b).
P2 expression in outer membrane preparations of wild-type and P2-deficient nontypeable Haemophilus influenzae (NTHI). (a) Proteins were resolved on a 12% acrylamide gel and stained with Coomassie Blue. Molecular mass standards are noted in kDa on the left. Arrows indicate presence of the P2 protein in the wild-type strain; (1) wild-type and (2) P2-deficient mutant. (b) Immunoblot assay of wild-type and P2-deficient mutants. Whole-cell lysate was resolved on a 12% acrylamide gel and transferred to nitrocellulose. Blots were probed with antiserum to its homologous P2. Bound antibodies were detected with anti-mouse IgG-horseradish peroxidase (HRP). Molecular mass markers are noted in kDa on the left; (1) wild type and (2) P2-deficient mutant.
Flow cytometry using P2-deficient mutants
To define the specificity of the immune response to P2, flow cytometry using the P2-deficient mutants was employed. Sera were tested at multiple dilutions and results are expressed for those dilutions that gave the optimal signal-to-noise ratio. For all three IN antisera tested, flow cytometry indicated a loss of surface binding in the P2 mutants in comparison to wild-type strains (Figure 5). These data support the conclusion that antibodies that bind to the surface of NTHI recognize epitopes on the P2 protein.
Assessment of serum antibodies induced by intranasal (IN) immunization by flow cytometry with wild-type and P2-deficient strains. Strains are noted along the X-axis. Results are expressed as the mean±s.e.m. Significance was determined with Student's t-test; WT: wild-type and ΔP2: P2-deficient mutant.
Cross-reactivity among strains
Whole-cell ELISA
To assess the extent to which antibodies to P2 bind to the surface of heterologous strains of NTHI, whole-cell ELISA was performed with 20 clinical isolates of NTHI. To facilitate screening this number of isolates and ensure availability of reagents, assays were performed with selected antisera: rP213 (20 μg IN), rP223 (50 μg IN), and rP254 (20 μg IN). These antisera were chosen as they demonstrated the highest degree of binding to the homologous strain in whole-cell ELISA. Binding of test sera was compared with that of normal mouse serum and Student's t-test was used to determine significance. Results from whole-cell ELISA indicate that anti-rP213 serum recognized 15 of 20 strains, anti-rP223 serum recognized 13 of 20 strains, and anti-rP254 recognized 15 of 20 strains (Figure 6). Substantial overlap was observed among the three sera with regard to strains that were recognized. However, some differences were noted. For example, 5P30H1 was the only strain to which anti-rP254 serum did not exhibit statistically significant binding, whereas both anti-rP213 and anti-rP223 sera displayed statistically significant binding to this strain, although this may be due to the aberrantly high standard deviation observed for this strain. From these experiments we conclude that antibodies to P2 induced by mucosal immunization recognize surface epitopes in multiple strains of NTHI in whole-cell ELISA.
Assessment of serum antibodies induced by intranasal (IN) immunization by whole-cell enzyme-linked immunosorbent assays (ELISAs) with heterologous strains of nontypeable Haemophilus influenzae (NTHI). Assays were performed in triplicate and results are expressed as the mean±s.e.m. Significance of antibody binding in comparison to normal mouse serum was determined using Student's t-test. IN, intranasal; OD, optical density; NMS, normal mouse serum. *P-value <0.05.
Flow cytometry
The same sera were also tested in flow cytometry with multiple strains. The sera were then tested in triplicate against 10–20 heterologous strains of NTHI, depending on availability of sera. By this method, anti-rP213 recognized 9 of 20 strains, anti-rP223 recognized 11 of 20 strains, and anti-rP254 recognized 5 of 10 strains (Figure 7). As in whole-cell ELISA, substantial overlap was observed among the strains testing positive, whereas some serum specificity was observed for other strains. Results of whole-cell ELISA and flow cytometry showed a degree of correlation with one another. However, a small number of strains that tested positive in ELISA were negative in flow cytometry, and vice versa. As both assays differ from each other in several respects, this is not unexpected. Overall, results of two independent methods indicate that antibodies to P2 recognize surface exposed epitopes on multiple strains of NTHI.
Assessment of serum antibodies induced by intranasal (IN) immunization by flow cytometry with heterologous strains of nontypeable Haemophilus influenzae (NTHI). Assays were performed in triplicate and results are expressed as the mean±s.e.m. Note that serum from mice immunized with rP254 was limited and was therefore only tested with 10 strains. Significance of antibody binding in comparison to normal mouse serum was determined using Student's t-test. IN, intranasal; NMS, normal mouse serum. *P-value <0.05.
Antibody avidity
The results of P2 ELISA, whole-cell ELISA, and flow cytometry revealed distinct differences in antibody populations generated by the two different routes of immunization. To further evaluate the differences between the antibodies induced by mucosal immunization compared with systemic immunization, the avidity of anti-rP2 antibodies was evaluated. Figure 8 shows that the avidity index of serum IgG antibodies for rP2 from SQ-immunized mice is approximately double that of serum IgG from IN-immunized mice. Although we cannot completely exclude the possibility that the sodium thiocyanate altered the conformation of P2 binding to the plate, these results support the conclusion that IN immunization yielded lower titer antibodies with reduced avidity that recognize surface exposed epitopes. In contrast, SQ immunization with rP2 in mice yielded high titer antibodies that exhibit high avidity but do not recognize surface epitopes.
Avidity of anti-recombinant P2 (rP2) antibodies induced by subcutaneous (SQ) vs. intranasal (IN) immunization. Avidity was tested by enzyme-linked immunosorbent assay (ELISA), using sodium thiocyanate (NaSCN) to disrupt antigen–antibody binding. Experiments were performed in triplicate. Results are expressed as the mean±s.e.m.
Discussion
As the most abundant protein in the outer membrane of NTHI, the major porin protein P2 was considered as a potential vaccine antigen for NTHI two decades ago. Antisera raised to killed bacteria that contained antibodies to P2 induced strain-specific passive protection in the chinchilla model of otitis media.28 Immunization of mice and rabbits with whole NTHI induce highly strain-specific antibodies to immunodominant, antigenically heterogeneous surface-exposed loops on the P2 protein.29 Natural infection in humans results in strain-specific, bactericidal antibodies to P2.30, 31 These lines of investigation suggested that the sequence heterogeneity of surface-exposed loops precluded P2 as a useful vaccine antigen. However, these studies all employed immunization with whole bacteria, which induces highly strain-specific responses to immunodominant surface loops of P2. In designing the present study, we speculated that immunization with purified P2 would induce a different set of antibodies to P2 compared with when the host is presented with whole bacterial cells. The present study is the first to assess immunization with full-length rP2 and to assess mucosal immunization with P2.
One reason we chose to evaluate both mucosal and systemic immunization is that NTHI is a respiratory pathogen. Although the relative importance of IgA antibodies in preventing infection with NTHI is not yet known, several studies provide evidence that mucosal IgA antibody is protective. Levels of secretory IgA antibody against P6 in breast-fed infants are inversely correlated with colonization of the nasopharynx with NTHI.23 Furthermore, intranasal immunization with rP4 and rP6 induces enhanced clearance of NTHI in animal models.26, 27, 32, 33
The mucosal immune system is now widely recognized to be compartmentalized. Intranasal immunization induces antibodies in the nasal, oral, respiratory, and vaginal secretions, whereas oral immunization preferentially induces antibodies within the gut.34 Immunization via the nasopharynx-associated lymphoid tissue induces both salivary and respiratory tract IgA responses.35, 36 For this reason, as well as several practical aspects, we chose to study mucosal IgA responses to rP2 in saliva as a surrogate for analyzing IgA responses within the lungs. In our study, intranasal immunization with P2 resulted in low levels of anti-P2 IgA in saliva. Whether these levels are physiologically significant is unknown, although it is anticipated that adjusting dosage and immunization schedules could improve mucosal responses.
Although titer is an important consideration, recognition of surface epitopes is also an important factor in antibody efficacy. For example, bactericidal or opsonizing antibodies bind to the bacterial surface. For this reason, flow cytometry and whole-cell ELISA were used to study binding of serum IgG to surface epitopes. In both assays, sera from IN-immunized animals recognized surface epitopes. In marked contrast, sera from SQ-immunized animals exhibited no binding to surface epitopes. This result has important implications. The observation emphasizes that the route of immunization has a profound effect on the characteristics of the resulting antibody response. Sera from SQ-immunized animals displayed a high titer of antibodies to rP2 in quantitative ELISA, but these antibodies appear to be largely directed toward buried epitopes. The reason for this difference is unclear, although we speculate that the difference may arise from differences in antigen processing due to the route of immunization or different modes of action of the two adjuvants used. Alternatively, it may be that emulsification with incomplete Freund's adjuvant partially denatured the rP2, and conformational epitopes necessary for inducing of antibodies to the surface of the organism were lost.
Another interesting observation was that in two instances, anti-rP213 (20 μg IN) and anti-rP254 (20 μg IN), sera that yielded low titers of anti-P2 serum IgG by quantitative ELISA (Table 1) displayed a high degree of recognition of surface epitopes by whole-cell ELISA and flow cytometry (Figures 2 and 3). Flow cytometry using mutants deficient in the P2 protein confirmed that these sera recognize surface epitopes on P2. As coating an ELISA plate with purified proteins is likely to distort some epitopes, particularly conformational epitopes, we hypothesize that our quantitative ELISAs underestimated the amount of anti-P2 serum IgG in these samples. This speculation is also supported by results of antibody avidity assays. Antisera from IN-immunized animals displayed lower avidity to rP2 when compared with antisera from SQ-immunized animals. This lower avidity may have contributed to the lower titers observed in antisera from mucosally immunized animals. It is important to note here that because the avidity experiments were performed with purified rP2, no conclusions can be drawn from this assay regarding the avidity of anti-P2 antibodies to epitopes on the bacterial surface as measured in whole-cell ELISA.
To determine the degree of cross-reactivity of anti-rP2 antibodies among strains, sera were tested for recognition of surface epitopes on 20 clinical isolates. Sera from intranasal immunization recognized 65–95% of strains by ELISA, and 25–55% of strains by flow cytometry. Overall, we conclude that antibodies to P2 induced by mucosal immunization recognize surface epitopes in multiple strains of NTHI. The cross-reactivity of antibodies from SQ-immunized animals, in contrast, could not be evaluated by whole-cell ELISA or flow cytometry as these antibodies did not recognize epitopes on the surface of the organism. Natural infection with NTHI induces highly strain-specific antibody responses to P2. Furthermore, immunization of animals with whole NTHI induces antibodies largely to one variable, immunodominant loop on P2.29 The results of the present study demonstrate that immunization with a purified form of the same protein induces a different set of antibodies compared with infection with whole NTHI. Mucosal immunization with purified P2 with cholera toxin as an adjuvant induced antibodies that recognize conserved epitopes present on multiple strains of the organism. This is an important finding, as it indicates that immunization with an antigen that contains both conserved and variable epitopes can induce antibodies to surface epitopes of multiple strains.
In summary, the present study shows that rP2 is immunogenic when administered both mucosally and systemically. However, marked differences were observed in the antibody populations produced by these two routes of immunization. When IN administered, rP2 induced antibodies in mice that recognize surface epitopes on the homologous strain, as well as several heterologous strains, implying a potential advantage for mucosal immunization over systemic immunization for rP2. Although future work is needed to assess the protective effect of immunization with rP2, our results indicate that P2 should be studied further as a vaccine candidate for NTHI.
Methods
Bacteria and growth conditions. Fifteen NTHI strains were isolated from sputum of adults enrolled in the chronic obstructive pulmonary disease Study Clinic at the Buffalo VA Medical Center 37 and eight strains were obtained by tympanocentesis of middle ear fluid from children with otitis media in various cities (Buffalo, Seattle, Dallas). NTHI was grown on chocolate agar plates or in brain heart infusion broth supplemented with 10 μg/ml hemin and 10 μg/ml β-nicotinamide adenine dinucleotide. Escherichia coli strains TOP 10 and BL21 DE3 pLysS (Invitrogen, Carlsbad, CA) were grown on Luria–Bertani plates, in Luria–Bertani broth, or in Terrific broth with appropriate antibiotics.
Cloning of ompP2 into pRSETB. The ompP2 gene was amplified by PCR from three strains of NTHI using previously described primers.38 Genomic DNA from strains 13P24H1, 54P24H1, and 23P2H1 were used as templates. The PCR products were digested with Bam HI (New England Biolabs, Ipswich, MA) and ligated into the vector pRSETB (Invitrogen), which places a 6X-Histidine tag on the N terminus of the recombinant protein. Ligated plasmid was transformed into chemically competent TOP 10 cells and positive clones were selected by plating onto Luria–Bertani plates containing carbenicillin. Colonies were subjected to PCR to identify insertion of the ompP2 gene. DNA sequencing (Roswell Park Biopolymer Facility, Buffalo, NY) was performed to ensure the gene was in frame and retained the correct sequence. The resulting plasmids were named pKO6 (strain 23P2H1), pKO7 (strain 54P24H1), and pKO10 (strain 13P24H1).
Expression and purification of rP2. Chemically competent BL21 DE3 pLysS cells were transformed with the plasmids pKO6, pKO7, and pKO10. Transformed colonies were used to inoculate 32 ml of Terrific Broth supplemented with carbenicillin and chloramphenicol. Cultures were incubated at 37 °C with shaking to OD600=0.400. The bacteria were centrifuged at 1,800×g for 20 min and resuspended in 200 ml fresh media containing carbenicillin. Isopropyl-β-D-thiogalactopyranoside was added to 1 mM and the cultures were incubated for 3 h at 37 °C with shaking. Expression of the rP2 protein was monitored by SDS-PAGE. The bacteria were centrifuged at 3,000×g for 20 min at 4 °C and the bacterial pellet was stored at −20 °C.
A modification of the protocol described by Singh et al. 39was used to purify and refold rP2. Bacterial pellets were thawed and resuspended in 20 ml phosphate-buffered saline (PBS) containing 0.1% Triton X-100 and EDTA-free complete protease inhibitor (Roche, Indianapolis, IN) and subjected to 3×30 s bursts of sonication with a Branson Sonifier 450 at 40% power. This was followed by centrifugation at 15,000×g for 20 min at 4 °C to isolate inclusion bodies. Inclusion bodies were solubilized in binding buffer (2 M urea, 20 mM sodium phosphate, 0.5 M NaCl, pH 12.5). Solubilized rP2 was then bound to ∼2 ml Ni+ Sepharose Fast Flow (GE Biosciences, Piscataway, NJ) overnight at 4 °C. The resin was poured into a column and the fall through was collected. The column was washed with several column volumes of binding buffer containing 15 mM imidazole. This was followed by elution of rP2 from the column with binding buffer containing 500 mM imidazole. Purified rP2 was then subjected to refolding by dilution. Protein was diluted slowly into 10 volumes of cold refolding buffer (2 M urea, 50 mM Tris-HCl, 10% glycerol, 5% sucrose, 0.5 mM EDTA, 0.05% Zwittergent 3–14, pH 8.0). This was followed by dilution with five volumes of refolding buffer containing no urea or Zwittergent. Finally, the protein was subjected to diafiltration with buffer composed of 50 mM Tris-HCl, 10% glycerol, 5% sucrose, 0.5 mM EDTA, 0.01% Zwittergent, pH 8.0 using an Amicon stirred cell apparatus and a 10,000 molecular weight cutoff filter. The solution was concentrated to ∼1 ml using an Amicon centrifugal filter device (10,000 molecular weight cutoff). Protein concentration was estimated using the Pierce BCA protein assay (Pierce Biotechnology, Rockford, IL), and protein integrity was monitored by SDS-PAGE. Purified rP2 was stored in aliquots at −20 °C.
Construction of P2 mutants. P2-deficient mutants of NTHI strains 13P24H1, 23P2H1, and 54P24H1 were constructed as previously described by Neary et al.21 Briefly, genomic DNA was prepared from a P2-deficient mutant for H. influenzae strain Rd in which the ompP2 gene had been replaced by a chloramphenicol cassette.21 Genomic DNA was restricted with Bgl II and incubated with a single colony of NTHI on chocolate agar for 5 h at 35 °C in 5% CO2. The bacterial film was then resusupended in 1 ml brain heart infusion and plated on chocolate agar containing chloramphenicol at 2 mg/ml. Plates were incubated at 35 °C in 5% CO2 and resulting colonies were subjected to PCR to identify insertion of the chloramphenicol cassette into ompP2. SDS-PAGE and immunoblotting confirmed absence of the P2 protein.
Isolation of OMPs for SDS-PAGE. Outer membrane preparations were prepared as previously described by Murphy et al.40 with minor modifications. NTHI was grown overnight on chocolate agar. The following day, bacteria were suspended in 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4. Samples were subjected to 3×10 s bursts of sonication at 40% power followed by centrifugation at 900×g to pellet unbroken cells. The supernatant was centrifuged at 13,000×g to pellet cell membranes. Membranes were solubilized with 1% N-lauroyl sarcosine in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer. OMPs were then pelleted by centrifugation at 13,000×g and resuspended in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer for SDS-PAGE analysis.
SDS-PAGE and immunoblot assays. Samples were run on a 12% acrylamide gel and stained with Coomassie Blue. For immunoblotting, antigens in gels were transferred onto a nitrocellulose membrane, blocked with 3% nonfat milk, and probed with antisera. Blots were developed with appropriate antibodies conjugated to horseradish peroxidase (HRP).
Intranasal immunization of animals. Groups of 10 BALB/c mice were immunized IN on days 0, 10, and 20. Groups received rP2 at doses of 20 or 50 μg of protein. Immunization was achieved by placement of up to 5 μl of immunogen near the nostril of an unanesthitized animal, which the animal then sniffed into its nostril. One μg of cholera toxin (Sigma, St Louis, MO) was administered with each dose as an adjuvant. On Day 26, 10 μg of carbachol in 100 μl was administered intraperitoneally to induce salivation and saliva was collected. On Day 28, mice were anesthetized with halothane and exsanguinated by retroorbital puncture. All animal studies were carried out in accordance with guidelines set by the Office of Laboratory Animal Welfare and the Western New York VA Institutional Animal Care and Use Committee.
Systemic immunization of animals. Groups of 10 BALB/c mice were immunized SQ with 20 μg of rP2 emulsified with an equal volume of incomplete Freund's adjuvant (Sigma). Immunizations were administered on days 0, 14, and 28. On Day 35, mice were anesthetized with halothane and exsanguinated by retroorbital puncture. Sera for each group of mice were pooled and stored in aliquots at −20 °C.
Quantitative ELISA. Antibody concentrations were determined by quantitative ELISA as described by Wu et al.36 The wells of a 96-well microtiter plate were coated with rP2. Samples were tested in duplicate in a series of twofold dilutions at an appropriate starting concentration and plates were incubated overnight. Nonfat dry milk (2%) in PBS served as both the blocking agent and diluent, except in the case of saliva, which was diluted in 10% bovine serum albumin in PBS. Antibodies were detected with anti-mouse IgG or IgA (KPL, Gaithersburg, MD) or anti-mouse IgM (Invitrogen) conjugated to HRP. To determine immunoglobulin concentrations, a set of standard wells was run simultaneously by coating wells with either anti-mouse IgG or IgA antibodies. Mouse Ig reference serum (MP Biomedicals, Solon, OH) was used to generate a standard curve for each plate using four parameter logistic algorithms (Microplate Manager III; BioRad, Hercules, CA). The standard curves were then used to interpolate concentrations of immunoglobulins in samples. Antibody levels in saliva were normalized in relation to total IgA levels to account for fluctuations in IgA levels due to salivary flow rate.
Whole-cell ELISA. NTHI was grown to mid-logarithmic phase, centrifuged at 1,800×g for 10 min, and resuspended in PBS to an OD600=0.200. The bacterial suspension was coated onto 96-well plates by incubation at 4 °C for 16 h. Wells were blocked with 2% Nonfat dry milk in PBS and sera was applied to the wells in duplicate at appropriate dilutions. To determine appropriate dilutions, initial experiments were performed at multiple twofold dilutions until a loss of signal was achieved, and dilutions that yielded optical densities in the linear portion of the curve were chosen for further analysis. Plates were incubated with sera for 16 h at room temperature. Antibodies were detected with anti-mouse IgG-HRP (KPL). Absorbances were read at 450 nm on a BioRad microtiter plate reader, and results were analyzed with Microplate Manager III software.
Flow cytometry. NTHI was grown to mid-logarithmic phase and 100 μl of culture was centrifuged at 4,000×g for 5 min. The bacterial pellet was resuspended in an appropriate dilution of serum in PBS. Appropriate dilutions were determined by performing initial experiments at multiple twofold dilutions until a loss of signal was achieved to identify the dilution of serum that yielded the greatest signal-to-noise ratio. Bacteria incubated in PBS alone were used as a negative control, and bacteria incubated with polyclonal antiserum from mice immunized with whole bacterial cells of the homologous strain of NTHI were used as a positive control. The sample was then incubated for 1 h at 37 °C. The bacteria were washed with PBS and resuspended in a 1:10 dilution of antibody conjugated to flourescein isothiocyanate (KPL). After 30 min of incubation at 37 °C, the sample was diluted with 900 μl PBS and fluorescence was detected with a FACSCalibur flow cytometer using Cell Quest 3.1 software (BD Biosciences, San Jose, CA).
Avidity ELISA. To assess the avidity of antibodies to rP2, ELISAs were performed as described by Vermont et al.41 Briefly, rP2 was coated onto a 96-well microtiter plate at 2 μg/ml. Sera to be tested were added at appropriate dilutions and incubated for 16 h at room temperature. Half of the wells on each plate were incubated with 1.5 M sodium thiocyanate for 15 min. The corresponding wells on the other half of the plate were incubated for an equal length of time with PBS as a control. Plates were washed and antibodies were detected with anti-mouse IgG-HRP. Results are expressed as an avidity index (AI), where AI=(OD450 with sodium thiocyanate)/(OD450 without sodium thiocyanate).
Statistical methods. All assays were performed in triplicate. Results of assays are expressed as the mean±s.e.m. and were analyzed by Student's t-test for unpaired samples. Calculations were made using Prism 4.0 for Macintosh (Graphpad Software; San Diego, CA).
Accession numbers. Nucleotide sequences for ompP2 of strains 13P24H1, 23P2H1, and 54P24H1 were previously determined by Hiltke et al.38 The respective GenBank accession numbers are AY051383, AY051393, and AY051401.
Disclosure
The authors declared no conflict of interest.
References
Sethi, S. & Murphy, T.F. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14, 336–363 (2001).
Murphy, T.F. & Sethi, S. Chronic obstructive pulmonary disease: role of bacteria and guide to antibacterial selection in the older patient. Drugs Aging 19, 761–775 (2002).
Murphy, T.F., Sethi, S. & Niederman, M.S. The role of bacteria in exacerbations of COPD. A constructive view. Chest 118, 204–209 (2000).
Klein, J.O. Otitis media. Clin. Infect. Dis. 19, 823–833 (1994).
St Geme, J.W. III The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19 (Suppl 1), S41–S50 (2000).
Bennett, K.E., Haggard, M.P., Silva, P.A. & Stewart, I.A. Behaviour and developmental effects of otitis media with effusion into the teens. Arch. Dis. Child 85, 91–95 (2001).
Pichichero, M.E. Pathogen shifts and changing cure rates for otitis media and tonsillopharyngitis. Clin. Pediatr. (Phila.) 45, 493–502 (2006).
Block, S.L. et al. Community-wide vaccination with the heptavalent pneumococcal conjugate significantly alters the microbiology of acute otitis media. Pediatr. Infect. Dis. J. 23, 829–833 (2004).
Casey, J.R. & Pichichero, M.E. Changes in frequency and pathogens causing acute otitis media in 1995–2003. Pediatr. Infect. Dis. J. 23, 824–828 (2004).
Tristram, S., Jacobs, M.R. & Appelbaum, P.C. Antimicrobial resistance in Haemophilus influenzae. Clin. Microbiol. Rev. 20, 368–389 (2007).
Brook, I., Yocum, P., Shah, K., Feldman, B. & Epstein, S. Increased antimicrobial resistance in organisms recovered from otitis media with effusion. J. Laryngol. Otol. 117, 449–453 (2003).
McMichael, J.C. & Green, B.A. Vaccines for Moraxella catarrhalis and non-typeable Haemophilus influenzae. Curr. Opin. Investig. Drugs 4, 953–958 (2003).
Poolman, J.T. et al. Developing a nontypeable Haemophilus influenzae (NTHi) vaccine. Vaccine 19 (Suppl 1), S108–S115 (2000).
Burns, J.L. & Smith, A.L. A major outer-membrane protein functions as a porin in Haemophilus influenzae. J. Gen. Microbiol. 133 (Pt 5), 1273–1277 (1987).
Bell, J., Grass, S., Jeanteur, D. & Munson, R.S. Jr Diversity of the P2 protein among nontypeable Haemophilus influenzae isolates. Infect. Immun. 62, 2639–2643 (1994).
Sikkema, D.J. & Murphy, T.F. Molecular analysis of the P2 porin protein of nontypeable Haemophilus influenzae. Infect. Immun. 60, 5204–5211 (1992).
Duim, B., Dankert, J., Jansen, H.M. & van Alphen, L. Genetic analysis of the diversity in outer membrane protein P2 of non-encapsulated Haemophilus influenzae. Microb. Pathog. 14, 451–462 (1993).
Forbes, K.J., Bruce, K.D., Ball, A. & Pennington, T.H. Variation in length and sequence of porin (ompP2) alleles of non-capsulate Haemophilus influenzae. Mol. Microbiol. 6, 2107–2112 (1992).
Cope, L.D., Pelzel, S.E., Latimer, J.L. & Hansen, E.J. Characterization of a mutant of Haemophilus influenzae type b lacking the P2 major outer membrane protein. Infect. Immun. 58, 3312–3318 (1990).
Neary, J.M., Yi, K., Karalus, R.J. & Murphy, T.F. Antibodies to loop 6 of the P2 porin protein of nontypeable Haemophilus influenzae are bactericidal against multiple strains. Infect. Immun. 69, 773–778 (2001).
Neary, J.M. & Murphy, T.F. Antibodies directed at a conserved motif in loop 6 of outer membrane protein P2 of nontypeable Haemophilus influenzae recognize multiple strains in immunoassays. FEMS Immunol. Med. Microbiol. 46, 251–261 (2006).
Kurono, Y. et al. Nasal immunization induces Haemophilus influenzae-specific Th1 and Th2 responses with mucosal IgA and systemic IgG antibodies for protective immunity. J. Infect. Dis. 180, 122–132 (1999).
Harabuchi, Y. et al. Human milk secretory IgA antibody to nontypeable Haemophilus influenzae: possible protective effects against nasopharyngeal colonization. J. Pediatr. 124, 193–198 (1994).
Wu, H.Y., Nahm, M.H., Guo, Y., Russell, M.W. & Briles, D.E. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with Streptococcus pneumoniae. J. Infect. Dis. 175, 839–846 (1997).
Sabirov, A., Kodama, S., Hirano, T., Suzuki, M. & Mogi, G. Intranasal immunization enhances clearance of nontypeable Haemophilus influenzae and reduces stimulation of tumor necrosis factor alpha production in the murine model of otitis media. Infect. Immun. 69, 2964–2971 (2001).
Sabirov, A., Kodama, S., Sabirova, N., Mogi, G. & Suzuki, M. Intranasal immunization with outer membrane protein P6 and cholera toxin induces specific sinus mucosal immunity and enhances sinus clearance of nontypeable Haemophilus influenzae. Vaccine 22, 3112–3121 (2004).
Hotomi, M. et al. A recombinant P4 protein of Haemophilus influenzae induces specific immune responses biologically active against nasopharyngeal colonization in mice after intranasal immunization. Vaccine 23, 1294–1300 (2005).
Barenkamp, S.J. Protection by serum antibodies in experimental nontypable Haemophilus influenzae otitis media. Infect. Immun. 52, 572–578 (1986).
Yi, K. & Murphy, T.F. Importance of an immunodominant surface-exposed loop on outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 65, 150–155 (1997).
Yi, K., Sethi, S. & Murphy, T.F. Human immune response to nontypeable Haemophilus influenzae in chronic bronchitis. J. Infect. Dis. 176, 1247–1252 (1997).
Sethi, S., Wrona, C., Grant, B.J. & Murphy, T.F. Strain-specific immune response to Haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 169, 448–453 (2004).
Bertot, G.M., Becker, P.D., Guzman, C.A. & Grinstein, S. Intranasal vaccination with recombinant P6 protein and adamantylamide dipeptide as mucosal adjuvant confers efficient protection against otitis media and lung infection by nontypeable Haemophilus influenzae. J. Infect. Dis. 189, 1304–1312 (2004).
DeMaria, T.F., Murwin, D.M. & Leake, E.R. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect. Immun. 64, 5187–5192 (1996).
Wu, H.Y. & Russell, M.W. Nasal lymphoid tissue, intranasal immunization, and compartmentalization of the common mucosal immune system. Immunol. Res. 16, 187–201 (1997).
Wu, H.Y., Nguyen, H.H. & Russell, M.W. Nasal lymphoid tissue (NALT) as a mucosal immune inductive site. Scand. J. Immunol. 46, 506–513 (1997).
Wu, H.Y. & Russell, M.W. Induction of mucosal immunity by intranasal application of a streptococcal surface protein antigen with the cholera toxin B subunit. Infect. Immun. 61, 314–322 (1993).
Sethi, S., Evans, N., Grant, B.J. & Murphy, T.F. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl. J. Med. 347, 465–471 (2002).
Hiltke, T.J., Sethi, S. & Murphy, T.F. Sequence stability of the gene encoding outer membrane protein P2 of nontypeable Haemophilus influenzae in the human respiratory tract. J. Infect. Dis. 185, 627–631 (2002).
Singh, S.M. & Panda, A.K. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 99, 303–310 (2005).
Murphy, T.F., Dudas, K.C., Mylotte, J.M. & Apicella, M.A. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 147, 838–846 (1983).
Vermont, C.L. et al. Antibody avidity and immunoglobulin G isotype distribution following immunization with a monovalent meningococcal B outer membrane vesicle vaccine. Infect. Immun. 70, 584–590 (2002).
Acknowledgements
This work was supported by NIH grants AI19641 (TFM) and DE06746 (MWR) and by the Department of Veterans Affairs. KLO was supported by NIH predoctoral training grant T32AI007614. We thank Xueya Cai for advice with statistical analysis, Rebecca Benz for animal technical assistance, and Danielle Addison for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ostberg, K., Russell, M. & Murphy, T. Mucosal immunization of mice with recombinant OMP P2 induces antibodies that bind to surface epitopes of multiple strains of nontypeable Haemophilus influenzae. Mucosal Immunol 2, 63–73 (2009). https://doi.org/10.1038/mi.2008.70
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2008.70
This article is cited by
-
Current and future prospects for a vaccine for nontypeable Haemophilus influenzae
Current Infectious Disease Reports (2009)