Abstract
Although short-stature homeobox-containing gene (SHOX ) haploinsufficiency is responsible for Léri–Weill dyschondrosteosis (LWD), the molecular defect has not been identified in ∼20% of Japanese LWD patients. Furthermore, although high prevalence of microdeletions affecting SHOX is primarily ascribed to the presence of repeat sequences such as Alu elements around SHOX, it remains to be determined whether microdeletions are actually mediated by repeat sequences. We performed multiple ligation probe amplification (MLPA) assay in six Japanese LWD patients with apparently normal SHOX, followed by fluorescent in situ hybridization (FISH) analysis and sequencing for polymerase chain reaction (PCR) products encompassing the deletion junctions in patients with abnormal MLPA patterns. Consequently, heterozygous intragenic deletions were identified in three cases, i.e., a 5,906-bp deletion involving exons 4–5 in case 1, a 5,594-bp deletion involving exons 4–6a in case 2, and a 50,199-bp deletion involving exons 4–6b in case 3. The deletion breakpoints of cases 1 and 2 were present in nonrepeat sequences, whereas those of case 3 resided within Alu elements. The results suggest that cryptic SHOX intragenic deletions account for a small fraction of LWD and that microdeletions affecting SHOX can be generated by repeat-sequence-mediated aberrant recombinations and by nonhomologous end joining.
Similar content being viewed by others
Introduction
Léri–Weill dyschondrosteosis (LWD) is a dominantly inherited skeletal dysplasia characterized by Madelung deformity and mesomelic short stature (Langer 1965). It is caused by haploinsufficiency of the short-stature homeobox-containing gene (SHOX) on the short arm pseudoautosomal region (PAR1) of the human sex chromosomes (Ogata 2002; Blaschke and Rappold 2006). To date, extensive studies have been performed, identifying multiple intragenic mutations (Niesler et al. 2007) and various submicroscopic deletions encompassing the entire SHOX coding region and/or the putative downstream enhancer region(s) (Kosho et al. 1999; Ogata 2002; Benito-Sanz et al. 2005, 2006a, b; Fukami et al. 2006; Huber et al. 2006; Sabherwal et al. 2007). Submicroscopic deletions are more frequent than intragenic mutations (Ogata 2002), and this would be consistent with repeat sequences being abundantly present around SHOX, because aberrant intrachromosomal or interchromosomal recombinations are prone to occur between such sequences (Ogata 2002; Blaschke and Rappold 2006). Indeed, Alu and L1 elements are abundant on the X chromosome, with Alu elements being more frequent and L1 elements being less frequent on the PAR1 than on the rest of the X chromosome (Lyon 2000; Blaschke and Rappold 2006). However, SHOX abnormalities have not been identified in a substantial fraction of LWD patients (Benito-Sanz et al. 2006a; Blaschke and Rappold 2006), and we have also failed to reveal SHOX abnormalities in ∼20% of Japanese LWD patients (Ogata 2002). Furthermore, it remains to be determined whether microdeletions around SHOX are directly mediated by repeat sequences.
Multiple ligation probe amplification (MLPA) is a recently developed method for relative quantification of single-copy sequences in the human genome (Schouten et al. 2002). It has been demonstrated as a powerful tool in the detection of deletions affecting several genes, including SHOX (Benito-Sanz et al. 2005, 2006a, b; Gatta et al. 2007). However, except for a single patient with a tiny deletion encompassing exons 4–6a of SHOX that could be revealed only by MLPA analysis (Benito-Sanz et al. 2006b), this method has been performed in patients with sex chromosomal abnormalities or relatively large deletions involving the entire SHOX coding region and/or the downstream enhancer region(s) that can be identified by other methods such as fluorescent in situ hybridization (FISH) analysis and microsatellite genotyping (Kosho et al. 1999; Benito-Sanz et al. 2006a, b; Gatta et al. 2007).
Here we report three cryptic SHOX intragenic deletions that were first identified by MLPA analysis. Characterization of deletions implies that microdeletions affecting SHOX can be generated by homologous and nonhomologous rearrangements.
Patients and methods
Patients
We studied six unrelated Japanese female patients (cases 1–6) with definitive LWD phenotype [Madelung deformity and mesomelic short stature ranging from −4.4 standard deviation (SD) to −2.0 SD] in whom SHOX abnormality was not demonstrated by direct sequencing of coding exons 2–6b, by FISH analysis with an ∼18-kb cosmid probe spanning from intron 2 to intron 6a (Kosho et al. 1999), and by microsatellite and single nucleotide polymorphism (SNP) genotyping for previously described multiple loci utilized for localizing downstream enhancer(s) (Benito-Sanz et al. 2005; Fukami et al. 2006; Huber et al. 2006).
MLPA analysis
This study was approved by the Institutional Review Board Committee at the National Center for Child Health and Development. After taking informed consent, MLPA was performed for cases 1–6 and three control female subjects using a SALSA MLPA Kit P018B (MRC-Holland, Amsterdam, the Netherlands) that contains probes for various parts of SHOX (SHOX-specific probes) (Fig. 1a) and multiple other loci (reference probes). The protocol was as described in the manufacturer’s instructions (Schouten et al. 2002). In brief, 50 ng of leukocyte genomic DNA was hybridized with the probe mix, and the hybridization mixture was subjected to ligase reaction and polymerase chain rection (PCR) amplification. Subsequently, the PCR products amplified from ligated probes were visualized on a 310 ABI PRISM genetic analyzer (ABI Prisms, Foster City, CA, USA). For each SHOX-specific probe, a relative peak area was calculated by dividing each measured peak area by the sum of peak areas of the reference probes. The relative areas were compared between cases and controls, and relative peak areas less than 65% of those of controls were assessed to be indicative of heterozygous deletions (Schouten et al. 2002; Kozak et al. 2006).
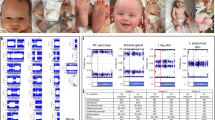
Identification of cryptic intragenic short-stature homeobox-containing gene ( SHOX) deletions in cases 1–3. a Summary of the multiple ligation probe amplification (MLPA) analyses and the fluorescent in situ hybridization (FISH) probes utilized in this study. The upper horizontal line indicates the physical distance from the Xp/Yp telomere. The rectangles represent SHOX exons (1–6b); the black and white areas denote the coding regions and the untranslated regions, respectively. The sites examined by MLPA probes (A–Q) are indicated by arrows; of these, the A–F sites reside on exons 1–6a, the H site lies just upstream of exon 6b, and thr I–Q sites reside around the enhancer region postulated at a position 30–250 kb downstream of SHOX coding region. The plus and the minus symbols represent the presence of two copies and a single copy of the corresponding sites, respectively. The lower thick lines represent the regions detected by FISH probes. The previously reported SHOX cosmid probe (Kosho et al. 1999) detects a ∼18-kb region from intron 2 to intron 6a, polymerase chain reaction (PCR) probe A detects a ∼4.0-kb region from intron 3 to intron 5, PCR probe B detects a ∼5.2-kb region just proximal to exon 6b, PCR probe C detects a ∼5.6-kb region at a position ∼19-kb proximal to exon 6b, and PCR probe D detects a ∼5.3-kb region at a position ∼32-kb proximal to exon 6b. The primers utilized were 5′-TCTCTCTCTGCTTCTCCCCA-3′ and 5′-GTGCAGGACGCGCGGT-3′ for PCR probe A, 5′-GTTAATGCTGAGAAGCTCTCCAAGCTAC-3′ and 5′-GTCCCTTACAAGGACACCTGTTATTGGAT-3′ for PCR probe B, 5′-GCTTGGTAGGAAGAGCCACAACTGTTCA-3′ and 5′-CTAGACGTCCACGGACCTATGTTGTAAC-3′ for PCR probe C, and 5′-TGAGTAATTAATCCCCACCAGTGAGGTC-3′ and 5′-CGTAGTTGGCCAAGACTCCACCATATTG-3′ for PCR probe D. b Results of MLPA assays (left part) and FISH analyses (right part). The peaks corresponding to sites around SHOX are indicated (A–Q); of these, the A–F sites reside on exons 1–6a, the H site lies just upstream of exon 6b, and I–Q sites reside around the enhancer region postulated at a position 30–250 kb downstream of SHOX coding region. MLPA assays indicate that compared with the relative peak areas in control female subjects, the relative peak areas for D and E in case 1, those for D–F in case 2, and those for D–H in case 3 are significantly reduced (shown in red). Relative peak areas for other loci (not specifically indicated) are similar between control female subjects and cases 1–3. The red peaks indicate the internal size markers. FISH analysis demonstrates that the ∼4-kb PCR probe A detects one faint and one clear signal in case 1 and only a single signal in cases 2 and 3, whereas the ∼18-kb cosmid probe detects two signals in cases 1–3
FISH analysis
FISH was performed with probes detecting the presumably deleted regions indicated by MLPA. The probes were obtained by long PCR using LA taq polymerase (Takara, Ohtsu) and were labeled with digoxigenin and detected by rhodamine antidigoxigenin. A SpectrumGreen-labeled probe for DXZ1 (Abbott, Abbott Park, IL, USA) was utilized as an internal control. For comparison, FISH was also performed with the cosmid probe (Fig. 1a) using previously described methods (Kosho et al. 1999).
Characterization of the deletions
Long PCR was performed with multiple primer pairs flanking the deleted regions. When long PCR products were obtained, they were subjected to direct sequencing using serial primers. The deletion size and the junction structure were determined by comparing the obtained sequences with the BX004827 and AL683871 sequences [National Center for Biotechnology Information (NCBI) database]. The presence or absence of repeat sequences around the breakpoints was examined with Repeatmasker (http://www.repeatmasker.org).
Results
MLPA analysis
Cryptic SHOX intragenic deletions were detected in cases 1–3 (Fig. 1b). Comparisons of relative peak areas indicated heterozygous deletion involving exons 4 and 5 in case 1, that involving exons 4–6a in case 2, and that involving exons 4–6b in case 3. No deletion was identified in cases 4–6. The results were reproduced in two independent experiments.
FISH analysis
FISH was performed with a PCR probe (A) for a region from intron 3 to intron 5 (Fig. 1a), detecting two signals with a marked difference in intensity (apparently one normal and one faint signal) in case 1 and only a single signal in cases 2 and 3 (Fig. 1b). In case 3 with a relatively large deletion, FISH was further carried out with three PCR probes (B, C, D), localizing the proximal breakpoint between the regions identified by PCR probes C and D (not shown). These microdeletions were not identified by the cosmid probe (Fig. 1b), as mentioned in “Patients”.
Deletion characterization
After examination with multiple primer sets, PCR products harboring the deletion junctions were obtained, and the deletion junction sequence was determined in cases 1–3 (Fig. 2a, b). Deletion size was 5,906-bp in case 1, 5,594-bp in case 2, and 50,199-bp in case 3. The deletion breakpoints of cases 1 and 2 were present on nonrepeat sequences, whereas those of case 3 resided within Alu elements. The fusion point resided at a 4-bp segment in case 1 and at a 30-bp segment in case 3 and was associated with an addition of an 8-bp segment of unknown origin in case 2.
Deletion sizes and junction structures. a Schematic representation of the intragenic deletions in cases 1–3. b Electrochromatograms showing the fusion points of the intragenic deletions. In case 1, a 4-bp segment surrounded by a rectangle is shared by the distal and the proximal breakpoint sequences. In case 2, an 8-bp segment of unknown origin surrounded by a rectangle is added to the deletion junction. In case 3, a 30-bp segment surrounded by a rectangle is shared by the distal and proximal breakpoint sequences. The breakpoints in cases 1 and 2 reside in nonrepeat sequences, whereas those in case 3 are present within Alu elements (shaded in light blue). Polymerase chain reaction (PCR) products encompassing the deletion junctions were obtained with the following primers: 5′-AAATTGGTTGTGGGGTGTGT-3′ and 5′-GTGCAGGACGCGCGG-3′ in case 1, 5′-TGGATCGTGAATCACTCCAA-3′ and 5′-GCCATCTCTACACCCGTGAT-3′ in case 2, and 5′-AACAGAGGTGAAGTGGATAATTGAG-3′ and 5′-TGGTTGGTTAGGAACTTGAATAGAG-3′ in case 3. The deletion junction sequences were determined with the following primers: 5′-AAATTGGTTGTGGGGTGTGT-3′in case 1, 5′-TTCGGTTCTCCTACAGGGTCT-3′ in case 2, and 5′-GCTAGGTGTGGTGGTGTGC-3′ in case 3. c Schematic representation of mechanisms leading to the generation of microdeletions. An aberrant intrachromosomal or interchromosomal recombination mediated by repeat sequences (green rectangles) causes a microdeletion (loss of a chromosomal region depicted by thin lines) between the repeat sequences. Nonhomologous end joining between nonrepeat sequences (a blue rectangle and a red ellipse) yields a microdeletion (thin lines) between the nonrepeat sequences and is often associated with an addition of a short segment of unknown origin (a green segment) or with a tiny overlapping segment common to the nonhomologous sequences (yellow segments)
Discussion
MLPA analysis identified cryptic SHOX intragenic deletions in cases 1–3. Since the cryptic deletions were detected in three of the six LWD patients with apparently normal SHOX, such tiny intragenic deletions may also be hidden in a substantial fraction of LWD patients without demonstrable SHOX haploinsufficiency. In this context, microdeletions affecting SHOX are frequently observed in LWD (Ogata 2002), and MLPA can identify at once various types of microdeletions affecting SHOX, including those involving a single or a few exons and those involving the entire coding region and/or the downstream enhancer region(s) (Benito-Sanz et al. 2006a, b), using genomic DNA of patients only. Thus, in conjunction with its simple and easy procedure, MLPA will serve as a powerful screening method for SHOX molecular defects.
The deletion junction resided in nonrepeat sequences in cases 1 and 2 and within Alu elements in case 3. The intragenic deletion in case 3 would be ascribed to an aberrant intrachromosomal or interchromosomal recombination mediated by repeat sequences (Fig. 2c) (Ogata 2002; Blaschke and Rappold 2006). By contrast, the intragenic deletions in cases 1 and 2 would be due to nonhomologous end joining (NHEJ), i.e., an aberrant breakage and re-union between nonhomologous sequences (Fig. 2c) (Shaw et al. 2004). In particular, the presence of a short segment of unknown origin at the deletion junction in case 2 is characteristic of NHEJ (Shaw et al. 2004). Furthermore, while a short segment common to distal and proximal breakpoint sequences was identified at the deletion junction in case 1, the segment appears to be too short to permit an aberrant recombination, and NHEJ associated with such a tiny overlapping segment has been reported previously (Kozak et al. 2006). In addition, the scattered distribution of the microdeletion breakpoints around SHOX (Kosho et al. 1999; Benito-Sanz et al. 2005, 2006a, b; Fukami et al. 2006; Huber et al. 2006; Sabherwal et al. 2007) may primarily reflect genomic rearrangements caused by NHEJ, and NHEJ may be facilitated by the high recombination frequency in the PAR1 and by the abundant presence of repeat sequences (e.g., Alu elements) (Shaw et al. 2004; Blaschke and Rappold 2006). Collectively, the present study implies that the microdeletions affecting SHOX can be caused by both homologous and nonhomologous rearrangements.
To date, we have examined a total of 29 families containing at least one patient with LWD and a normal karyotype (total 50 patients). Consequently, we identified various types of SHOX abnormalities in 26 of the 29 families (∼90%), i.e., 12 microdeletions involving the entire coding region and the putative downstream enhancer region(s), three microdeletions involving the entire coding region alone, three microdeletions involving the enhancer region(s) alone, three intragenic deletions, and five intragenic mutations (Kosho et al. 1999; Ogata 2002; Fukami et al. 2006; unpublished data) (Fig. 3). The frequency of SHOX abnormalities is higher than that reported by other groups (50–90%) (Blaschke and Rappold 2006). In particular, Benito-Sanz et al. (2006a) detected SHOX abnormalities only in 16 of 26 Spanish probands after performing extensive analysis, including MLPA analysis. Although the cause of the difference in the frequency of SHOX abnormalities (especially deletions) remains to be examined, this could be due to ethnic differences. Indeed, deletions encompassing NSD1 for Sotos syndrome are also much more frequently identified in Japanese than in other ethnic groups (Kurotaki et al. 2003).
Short-stature homeobox-containing gene (SHOX) abnormalities in 29 Japanese families with Léri–Weill dyschondrosteosis (LWD) and a normal karyotype. The upper horizontal line indicates the physical distance from the Xp/Yp telomere (“Tel”). The rectangles represent SHOX coding region. The black arrows denote the deleted regions. SHOX molecular defects were not identified in three families
SHOX molecular defects has not been demonstrated so far in cases 4–6. There are two possible explanations for this. First, there still may be a hidden abnormality impairing SHOX. For example, a mutation or a tiny deletion may exist in the promoter region, the nonexamined exonic sequences (MLPA examines only a part of exonic sequences), the intronic sequences, or the enhancer sequences. Second, there may be a mutation in some gene(s) other than SHOX. In this regard, one possible locus may reside near the HOXD gene cluster. An association between LWD-like skeletal abnormality and a balanced translocation t(2;8)(q31;p21) has been found in a father and his three children, with the 2q31 breakpoint being mapped near the HOXD gene cluster (Spitz et al. 2002), and chromosomal breakage around the HOXD cluster is known to result in various limb malformations (Dlugaszewska et al. 2006).
In summary, the results suggest that MLPA analysis is a highly useful method to identify microdeletions affecting SHOX, including cryptic intragenic deletions, and that such microdeletions can be caused by homologous sequence-mediated aberrant recombinations and by nonhomologous end joining. Further studies will permit re-evaluation of the prevalence of SHOX molecular defects and the mechanisms leading to microdeletions affecting SHOX.
References
Benito-Sanz S, Thomas NS, Huber C, Gorbenko del Blanco D, Aza-Carmona M, Crolla JA, Maloney V, Rappold G, Argente J, Campos-Barros A, Cormier-Daire V, Heath KE (2005) A novel class of Pseudoautosomal region 1 deletions downstream of SHOX is associated with Leri–Weill dyschondrosteosis. Am J Hum Genet 77:533–544
Benito-Sanz S, del Blanco DG, Aza-Carmona M, Magano LF, Lapunzina P, Argente J, Campos-Barros A, Heath KE (2006a) PAR1 deletions downstream of SHOX are the most frequent defect in a Spanish cohort of Leri–Weill dyschondrosteosis (LWD) probands. Hum Mutat 27:1062
Benito-Sanz S, Gorbenko del Blanco D, Huber C, Thomas NS, Aza-Carmona M, Bunyan D, Maloney V, Argente J, Cormier-Daire V, Campos-Barros A, Heath KE (2006b) Characterization of SHOX deletions in Leri–Weill dyschondrosteosis (LWD) reveals genetic heterogeneity and no recombination hotspots. Am J Hum Genet 79:409–414
Blaschke RJ, Rappold G (2006) The pseudoautosomal regions, SHOX and disease. Curr Opin Genet Dev 16:233–239
Dlugaszewska B, Silahtaroglu A, Menzel C, Kubart S, Cohen M, Mundlos S, Tumer Z, Kjaer K, Friedrich U, Ropers HH, Tommerup N, Neitzel H, Kalscheuer VM (2006) Breakpoints around the HOXD cluster result in various limb malformations. J Med Genet 43:111–118
Fukami M, Kato F, Tajima T, Yokoya S, Ogata T (2006) Transactivation function of an approximately 800-bp evolutionarily conserved sequence at the SHOX 3′ region: implication for the downstream enhancer. Am J Hum Genet 78:167–170
Gatta V, Antonucci I, Morizio E, Palka C, Fischetto R, Mokini V, Tumini S, Calabrese G, Stuppia L (2007) Identification and characterization of different SHOX gene deletions in patients with Leri–Weill dyschondrosteosys by MLPA assay. J Hum Genet 52:21–27
Huber C, Rosilio M, Munnich A, Cormier-Daire V, French SHOX GeNeSIS Module (2006) High incidence of SHOX anomalies in individuals with short stature. J Med Genet 43:735–739
Kosho T, Muroya K, Nagai T, Fujimoto M, Yokoya S, Sakamoto H, Hirano T, Terasaki H, Ohashi H, Nishimura G, Sato S, Matsuo N, Ogata T (1999) Skeletal features and growth patterns in 14 patients with haploinsufficiency of SHOX: implications for the development of Turner syndrome. J Clin Endocrinol Metab 84:4613–4621
Kozak L, Hrabincova E, Kintr J, Horky O, Zapletalova P, Blahakova I, Mejstrik P, Prochazkova D (2006) Identification and characterization of large deletions in the phenylalanine hydroxylase (PAH) gene by MLPA: evidence for both homologous and non-homologous mechanisms of rearrangement. Mol Genet Metab 89:300–309
Kurotaki N, Harada N, Shimokawa O, Miyake N, Kawame H, Uetake K, Makita Y, Kondoh T, Ogata T, Hasegawa T, Nagai T, Ozaki T, Touyama M, Shenhav R, Ohashi H, Medne L, Shiihara T, Ohtsu S, Kato Z, Okamoto N, Nishimoto J, Lev D, Miyoshi Y, Ishikiriyama S, Sonoda T, Sakazume S, Fukushima Y, Kurosawa K, Cheng JF, Yoshiura K, Ohta T, Kishino T, Niikawa N, Matsumoto N (2003) Fifty microdeletions among 112 cases of Sotos syndrome: low copy repeats possibly mediate the common deletion. Hum Mutat 22:378–387
Langer LO Jr (1965) Dyschondrosteosis, a heritable bone dysplasia with characteristic roentgenographic features. AJR Am J Roentgenol 95:178–188
Lyon MF (2000) LINE-1 elements and X chromosome inactivation: a function for “junk” DNA? Proc Natl Acad Sci USA 97:6248–6249
Niesler B, Röth R, Wilke S, Fujimura F, Fischer C, Rappold G (2007) The novel Human SHOX allelic variant database. Hum Mutat 28:933–938
Ogata T (2002) SHOX haploinsufficiency: lessons from clinical studies. Curr Opin Endocrinol Diabetes 9:13–20
Sabherwal N, Bangs F, Roth R, Weiss B, Jantz K, Tiecke E, Hinkel GK, Spaich C, Hauffa BP, van der Kamp H, Kapeller J, Tickle C, Rappold G (2007) Long-range conserved non-coding SHOX sequences regulate expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum Mol Genet 16:210–222
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30:e57
Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13(Spec No 1):R57–R64
Spitz F, Montavon T, Monso-Hinard C, Morris M, Ventruto ML, Antonarakis S, Ventruto V, Duboule D (2002) A t(2;8) balanced translocation with breakpoints near the human HOXD complex causes mesomelic dysplasia and vertebral defects. Genomics 79:493–498
Acknowledgments
We thank Professor Gudrun Rappold, Heidelberg University, for her critical comments, and Mr. Shunji Yamamori, Mitsubishi Chemical Medience Corporation, Tokyo, Japan, for his technical assistance in FISH analysis. This study was supported by Grants for Child Health and Development (17C-2) and for Research on Children and Families (H18-005) from the Ministry of Health, Labor, and Welfare, and by Grants-in-Aid for Scientific Research (category C: 18591178) from the Ministry of Education, Culture, Sports, Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fukami, M., Dateki, S., Kato, F. et al. Identification and characterization of cryptic SHOX intragenic deletions in three Japanese patients with Léri–Weill dyschondrosteosis. J Hum Genet 53, 454–459 (2008). https://doi.org/10.1007/s10038-008-0269-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-008-0269-z
Keywords
This article is cited by
-
Identification of 15 novel partial SHOX deletions and 13 partial duplications, and a review of the literature reveals intron 3 to be a hotspot region
Journal of Human Genetics (2017)
-
Identification of a novel 15.5 kb SHOX deletion associated with marked intrafamilial phenotypic variability and analysis of its molecular origin
Journal of Genetics (2016)
-
Polymorphism of 3′ UTR of MAMLD1 gene is also associated with increased risk of isolated hypospadias in Indian children: a preliminary report
Pediatric Surgery International (2016)
-
Rare pseudoautosomal copy-number variations involving SHOX and/or its flanking regions in individuals with and without short stature
Journal of Human Genetics (2015)
-
Spectrum of phenotypic anomalies in four families with deletion of the SHOX enhancer region
BMC Medical Genetics (2014)