Abstract
Spinocerebellar ataxia type 1 (SCA1; OMIM: #164400) is an autosomal dominant cerebellar ataxia caused by an expansion of CAG repeat, which encodes polyglutamine, in the ataxin-1 (ATXN1) gene. Length of polyglutamine in the ATXN1 protein is the critical determinant of pathogenesis of this disease. Molecular diagnosis of SCA1 is usually undertaken by assessing the length of CAG repeat configuration using primers spanning this configuration. However, this conventional method may potentially lead to misdiagnosis in assessing polyglutamine-encoding CAG repeat length, since CAT interruptions may be present within the CAG repeat configuration, not only in normal controls but also in neurologically symptomatic subjects. We developed a new method for assessing actual CAG repeat numbers not interrupted by CAT sequences. Polymerase chain reaction using a primer pair labeled with two different fluorescences followed by restriction enzyme digestion with SfaNI which recognizes the sequence “GCATC(N)5”, lengths of actual CAG repeats that encode polyglutamine were directly detected. We named this method “dual fluorescence labeled PCR-restriction fragment length analysis”. We found that numbers of actual CAG repeat encoding polyglutamine do not overlap between our cohorts of normal chromosomes (n = 385) and SCA1 chromosomes (n = 5). We conclude that the present method is a useful way for molecular diagnosis of SCA1.
Similar content being viewed by others
Introduction
Spinocerebellar ataxia type 1 (SCA1) (OMIM: #164400) is caused by an expansion of trinucleotide (CAG) repeat that encodes polyglutamine tract in the ataxin-1 (ATXN1) gene (OMIM: *601556) lying in the short arm of human chromosome 6 (6p23) (Chung et al. 1993; Orr et al. 1993; Banfi et al. 1994). In normal chromosomes, this CAG repeat shows repeat-length polymorphism ranging in size between 19 and 39 repeats. In contrast, the length of expanded CAG repeat in the SCA1 disease chromosomes ranges from 39 up to 81 repeats. The length of expansion is inversely correlated with age-of-onset of disease, suggesting a direct role of CAG repeat/polyglutamine length in the pathogenesis of SCA1 (Chung et al. 1993; Orr et al. 1993; Banfi et al. 1994).
The ataxin-1 (ATXN-1) gene encodes a protein called ataxin-1 (ATXN-1), which is a 816-amino acid protein with a molecular mass of 87 kDa. The polyglutamine tract lies at its amino (N)-terminal region. In addition to the polyglutamine tract, there are some other important domains in ATXN-1, such as nuclear localization signal and the AXH domains. Although the functions of ATXN-1 are not fully understood, recent investigation suggests that the AXH domain in the ATXN-1 interacts with Gfi-1/Senseless protein depending on the length of the polyglutamine tract, resulting in reduction of the Gfi-1 level, which in turn may contribute to neurodegeneration (Tsuda et al. 2005).
The molecular diagnosis of SCA1 is usually undertaken by PCR amplification using primer pairs spanning the CAG repeat (Matilla et al. 1993; Orr et al. 1993; Goldfarb et al.1996). However, this conventional method may contain caveats mainly stemming from the presence of trinucleotide CAT interruption sequence(s) within the CAG repeat tract. Most (98%) of the normal alleles contain one to three CAT interruptions, resulting in a much shorter polyglutamine tract when it is translated. On the other hand, expanded CAG repeats are characterized by continuous, “pure” CAG stretches, resulting in pure polyglutamine expansions (Chung et al. 1993). This fact would imply that the diagnosis of SCA1 is important to assess, not merely by the length of “CAG repeat configuration” (i.e., total length of CAG and CAT repeats), but by determining the length of pure CAG repeat encoding polyglutamine. Indeed, some exceptional cases have been previously reported. An allele with 44 repeats, which indicates “expansion” in conventional method, has been reported in an asymptomatic subject (Quan et al. 1995). This subject harbored CAG repeat configuration of (CAG)12CATCAGCAT (CAG)12CATCAGCAT (CAG)14, showing that this allele encodes for polyglutamine tract with normal length due to four CAT interruptions. Another example has been reported in an expanded allele with 58 repeats interrupted with two CAT sequences as, 5′-(CAG)45CATCAGCAT(CAG)10-3′ (Matsuyama et al. 1999). Particularly, special caution would be needed when assessing the CAG repeat length near the border of normal and expanded repeats, since the upper limit of normal repeat and the shortest expansion overlap at 38 repeats. While “pure” 38 CAG stretch is pathogenic, the 38 CAG repeat with CAT interruption is not pathogenic due to the much shorter polyglutamine stretch (Ranum et al. 1994). From these observations, it would be better if one could directly assess the actual CAG repeat length not interrupted by CAT sequences, since the length of polyglutamine tract in the ATXN-1 is the only basic defect that leads to pathogenesis.
In this study, we developed a new method that would allow one to detect the actual number of CAG repeat length not interrupted by CAT sequence(s). We show here that, by using our new method, one can directly assess 5′- and 3′- CAG repeat numbers disrupted by CAT sequence(s) which may be contained in the CAG repeat configuration. We propose that this new method, “dual fluorescence labeled PCR-restriction fragment length analysis”, is a direct way to accurately diagnose SCA1. We not only introduce this new method, but we also show that actual “pure” CAG repeat numbers do not overlap between normal Japanese and SCA1 subjects by using this method. We also describe a unique family with short, but pathogenic, CAG repeats detected with our new method.
Materials and methods
Overall design of the dual fluorescence labeled PCR-restriction fragment length analysis
As previously described by others (Chung et al. 1993), every CAT interruption within “the CAG repeat configuration” (i.e., total length of CAG and CAT repeats) is theoretically recognized and digested with the restriction enzyme, “SfaNI”, which recognizes 5′-GCATC(N)5-3′. There are no other consensus SfaNI sites outside the CAG repeat configuration when the genome is amplified using appropriate primers. Using this advantage, we designed a comparison of fragment lengths of the PCR product both with and without the SfaNI digestion (Fig. 1).
A basic concept of the dual-fluorescence labeled PCR-restriction fragment length analysis. T Length of PCR product containing whole CAG repeat configuration. S Fragment length of 5′ sense-strand PCR product, labeled by FITC (fluorescence isothiocyanate) yielded after restriction enzyme SfaNI, digestion. AS Fragment length of 3′ anti-sense-strand PCR product labeled by VIC yielded after restriction enzyme SfaNI, digestion. A In case of no CAT interruption in the CAG repeat configuration, PCR fragment without SfaNI (T), 5′ sense-strand PCR product labeled by FITC yielded after SfaNI digestion (S), and the 3′ anti-sense-strand PCR product labeled by VIC yielded after SfaNI digestion (AS) all coincide in length on ABI PRISM 3100-Avant System. B In case one, CAT interruption is present within the CAG repeat configuration. The length of PCR fragment without SfaNI (T) is calculated as the sum of (5′ sense-strand PCR product labeled by FITC yielded after SfaNI digestion: S), (3′ anti-sense-strand PCR product labeled by VIC yielded after SfaNI digestion: AS), and 3 base-pair (bp) due to trinucleotide, GTC, on anti-sense strand (underlined). T = S + AS + 3 (bp). C In case two, CAT interruptions are present within the CAG repeat configuration. Two different SfaNI-digestion sites (a and b) are predicted (right panel). Theoretically, four types of fragments would be yielded due to combination of complete and partial digestions (two FITC-labeled fragments S1; ad S2, and two VIC-labeled fragments, AS1 and AS2). The length of PCR fragment without SfaNI (T) could be calculated as the sum of S1 + AS2 + 9 bp (corresponding nine-nucleotide “GTCGTCGTC”, underlined in right panel)
When there are no CAT interruptions within the CAG repeat configuration, the fragment length after SfaNI digestion will theoretically be the same as that without digestion (Fig. 1A).
When there is one CAT interruption, the length of the PCR product without the digestion would be calculated as the sum of lengths of two different fragments yielded after the digestion: the sum of the lengths of “the 5′-digested fragment”, “the 3′-digested fragment”, and 3 (Fig. 1B). The 5′- and 3′- SfaNI digested fragments could be differentiated if the sense and anti-sense primers are labeled with different fluorescent dyes.
When there are two CAT interruptions, the length of the PCR product without the digestion would be calculated as the sum of lengths of three different fragments yielded after the digestion: “5′-digested fragment”, “GCATC(N)5”, and “the 3′-digested fragment” (Fig. 1C). Conversely, the size difference between fragment length without the digestion and the sum of “5′-digested fragment” and “the 3′-digested fragment”, would be the length of “GCATC(N)5”.
CAG repeat configurations containing more than two CAT interruptions were not seen in our cohort of samples (data shown in Results). We therefore artificially generated such clones using site-directed mutagenesis (Invitrogen, Calif., USA). When there are more than two CAT interruptions, the length of PCR fragments without the enzyme digestion “minus” the sum of lengths of the 5′- and 3′- SfaNI digested fragments would be the differences harboring CAT interruptions.
Thus, it seemed theoretically possible to detect both 5′- and 3′-CAG repeat sequences disrupted by CAT interruption. We tested our hypothesis first by examining 10 plasmid clones containing ATXN1 CAG repeat configurations obtained from five individuals (Table 1).
DNA samples and materials
One hundred and ninety-one control individuals and two SCA1 subjects were first analyzed. The control group comprised of 60 neurologically normal subjects, 50 subjects with cerebrovascular diseases, in whom no family history of ataxia was present, and 81 individuals with dominantly inherited ataxias previously excluded for SCA1 by conventional diagnostic method. Two subjects with typical clinical features of SCA1 had been molecularly confirmed as having this disease by the conventional method. Peripheral blood samples were obtained after informed consent, and DNA was extracted as reported elsewhere (Ishikawa et al. 1997). The study was approved by the Institutional Review Board of Tokyo Medical and Dental University.
Detection of CAG repeat/CAT interruption with automated fluorescence sequencer
For amplifying the CAG repeat configuration, a primer set “CAG-a and CAG-b” (Chung et al.1993) was used, although the sense primer was labeled with FITC (fluorescent isothiocyanate) at its 5′-end (CAG-b: 5′-[FITC] CCAGACGCCGGGACACAAGGCTGAG-3′), and the anti-sense primer was 5′-end labeled with VIC (CAG-a: 5′-[VIC]-CCGGAGCCCTGCTGAGGTG-3′). PCR was performed in an ordinary condition in a final volume of 25 μl, containing 50 ng of genomic DNA, 4.0 pmol of each primer, 1 μl of 10% dimethylsulfoxide (DMSO), 2 mM each deoxynucleosides (dNTPs), and 1.0 unit of Gold Tag DNA polymerase (Takara, Japan). Thermal setting was initial denature at 95°C for 4 min, 3 cycles of 95°C for 1 min, 70°C for 30 s, 72°C for 30 s; subsequent 3 cycles of 95°C for 1 min, 68°C for 30 s, 72°C for 30 s; 3 cycles of 95°C for 1 min, 66°C for 30 s, 72°C for 30 s; 3 cycles of 95°C for 1 min, 64°C for 30 s, 72°C for 30 s; final 20 cycles of 95°C for 1 min, 62°C for 30 s, 72°C for 30 s, and final extension at 72°C for 10 min. For each reaction, 10 μl of PCR product was digested with SfaNI (New England BioLabs, USA) in a final volume of 15 μl containing optimal reaction condition (10 mM sodium chloride, 5 mM Tris–hydrochloride, 1 mM Mg Cl2, 0.1 mM dithiothreitol (DTT; pH 7.9) incubated at 37°C, as recommended by the supplier (Chung et al. 1993). PCR products both with and without the SfaNI digestion were diluted with the same solution (Hi-Di Formamide and Gene Scan-500 LIZ Size Standard), and then heated at 95°C for 2 min for denature, immediately cooled within ice, and loaded on ABI PRISM 3100-Avant System (Applied Biosystems). The electrophoresis was undertaken at 50°C, with 15 volts.
Evaluation of dual fluorescence labeled PCR-restriction fragment length analysis
We first randomly chose five individuals with different CAG repeat configurations (Table 1). On each subject, genomic DNA was amplified with CAG-a and -b primers, and PCR product was subcloned into PCR-TOPO (Invitrogen, USA). Then, 10 clones from each individual were sequenced with universal and reverse primers as previously described (Li et al. 2003). These five individuals harbored any of the CAG repeat configurations without CAT interruption, or 1–2 CAT interruption(s). Then, the dual fluorescent-labeled PCR-restriction fragment length analysis was performed on these clones to check the consistency.
Results
Evaluation of dual fluorescence labeled PCR-restriction fragment length analysis
To evaluate consistency of our new method, we first examined on 10 plasmid clones which contained CAG repeat configuration in the ATXN1 gene (Table 1). On the plasmid clones without CAT interruption within the CAG repeat configuration (e.g., Case 1, Allele 1), there were no differences in fragment lengths on ABI 3100-Avant System between data with and without the SfaNI restriction enzyme digestion (Fig. 1A), as has been hypothesized.
When plasmids with one CAT interruption were examined (e.g., Table 1, Case 2, Allele 2), the fragment length of the PCR product amplified by the two primers should be the sum of the lengths of “5′-digested fragment labeled with FITC”, that of “3′-digested fragment labeled with VIC”, and 3. The SfaNI digestion yielded was two fragments exactly corresponding the 5′-digested fragment labeled with FITC and the 3′-digested fragment labeled with VIC (Fig. 1B).
If the CAG repeat configuration contained two CAT interruptions, the PCR product should be separated into three fragments: “5′-digested fragment labeled with FITC”, “3′-digested fragment labeled with VIC”, and the “internal fragment limited by the two SfaNI-recognition sites”. Since this internal fragment is not labeled, this fragment will not be detected. However, the length of the “internal fragment limited by the two SfaNI-recognition sites” could be calculated by the following formula: (the length of internal sequence between the two CAT interruptions) = (the length of PCR product without SfaNI digestion) – (the sum of lengths of “5′-digested fragment labeled with FITC” and “3′-digested fragment labeled with VIC”). This was indeed confirmed by the experiment on plasmids with two CAT interruptions (e.g., Table 1, Case 4, Allele 1) with the dual fluorescence labeled PCR-restriction fragment length analysis (Fig. 1C).
These examinations not only confirmed our hypothesis, but also suggested that the new method is able to directly assess nucleotide sequences of CAG repeat configuration. If the length-difference between the fragments yielded with and without the SfaNI restriction enzyme digestion was nine nucleotide long, the nucleotide excised by the SfaNI was always as “5′-CATCAGCAT-3′”, confirmed by experiments on plasmid clones. Similarly, if the length-difference was 12 nucleotides, the internal sequence was always as “5′-CAT(CAG)2CAT-3′”. If the lengths was 15 nucleotides, the internal sequence was always as “5′-CAT(CAG)3CAT-3′”.
We did not find DNA samples with more than two CAT interruptions with the CAG repeat configuration in our cohort of 191 control subjects (data described later). To check the usefulness of our new method for alleles with more than two CAT interruptions, a CAG repeat configuration with three CAT interruptions was artificially generated from a plasmid clone, Case 5 Allele 2 (Table 1), by using site-directed mutagenesis. The exact sequence of this configuration containing three CAT interruption was “5′-(CAG)9CAT(CAG)6CATCAGCAT(CAG)10-3′”. When digested with SfaNI, three fragments corresponding 5′-completely digested fragment labeled with FITC, 3′-completely digested fragment labeled with VIC, and 3′-partially digested fragment labeled with VIC, were detected (Fig. 2). The combined length of the 5′-completely digested fragment labeled with FITC (S1) and the 3′-completely digested fragment labeled with VIC (AS2) was 18 base-pairs (bp) shorter than the length of total CAG repeat configuration, consistent with the fact that six CAG repeats were lying between two CAT interruption sequences.
The electrophoresis pattern of the dual-fluorescence labeled PCR-restriction fragment length analysis on a plasmid clone containing three CAT interruptions. A The fragment analysis on ABI 3100 without SfaNI digestion. FITC-labeled PCR product (arrow #1) and VIC-labeled PCR product (arrow #2) which are actually the same size in length, differ in size for 5.8 base-pairs (bp) on this analyzing system. This 5.8-bp gap between the FITC and VIC fragments was always constantly seen on ABI 3100. B The fragment analysis after SfaNI digestion. Three major peaks corresponding to a completely digested VIC-labeled 3′ anti-sense-strand PCR fragment (arrow #3; designated, AS2), a partially digested, VIC-labeled 3′ anti-sense-strand PCR fragment (arrow #4; designated, AS1), and a completely digested, FITC-labeled 5′ sense-strand PCR product (arrow #5; designated, S1), are demonstrated. When a total length of PCR fragment containing whole CAG repeat configuration is designated “T”, T = AS1 + AS2 + S1 + 18 (bp) was confirmed both by fragment length analysis and actual sequence analysis. Although presence of a partially digested FITC-labeled 5′ anti-sense-strand PCR fragment (designated, S2) has been considered on hypothesis, it was never observed on ABI3100 sequencer. Therefore, we considered that FITC-labeled PCR product directly reflects a completely digested fragment (S1)
Based on these observations, we confirmed that the dual fluorescence labeled PCR-restriction fragment length analysis appeared to be a useful and rapid way to directly assess CAG repeat configuration.
Results on 191 control individuals and two clinically typical SCA1 subjects
We next examined 191 control Japanese individuals and two SCA1 subjects by the dual fluorescence labeled PCR-restriction fragment length analysis. The two most frequent CAG repeat configurations were those with 26 and 28 combined CAG and CAT repeats (Fig. 3). The frequencies of these alleles in normal Japanese chromosomes were 27.1% for 28 CAG/CAT repeat-units, and 24.0% for 26 CAG/CAT repeat-units. The range of CAG repeat configuration was from 17 up to 40 repeat-units in control groups. Two SCA1 subjects harbored 46 and 54 repeat-units on their SCA1 chromosomes.
The analysis of the SCA1 CAG repeats in control Japanese individuals. A The distribution of CAG repeat configuration (i.e., combined CAG repeat and CAT interruptions). In the present cohort of 193 Japanese subjects, the CAG repeat number which may contain CAT interruptions ranged from 17 to 40 repeat-unit. B The distribution of actual (pure) CAG repeat number encoding polyglutamine in the region 5′ (upstream) of the first CAT interruption. Pure CAG repeat in the 5′-region ranges from 10 to 20 repeat-units, and two major peaks are seen at 11 and 13 CAG repeats. Notice that a chromosome with 27 pure CAG repeat was an exceptionally large repeat. C The distribution of actual (pure) CAG repeat number encoding polyglutamine in the region 3′ (downstream) of the last CAT interruption. Pure CAG repeat in the 3′-region ranges from 7 to 23 repeat-units, and two major peaks are seen at 10 and 16 CAG repeats
When 386 chromosomes from 193 individuals were examined by the dual fluorescence labeled PCR-restriction fragment length analysis, we found that 140 chromosomes had one CAT interruption with the CAG repeat configuration, counting 36.4% of our cohort of Japanese control chromosomes (Table 2). On the other hand, there were 243 chromosomes with two CAT interruptions, counting 62.9% of 386 chromosomes. Table 3 shows exact sequences of CAG repeat configurations that contain either 1 or 2 CAT interruption found in this study. There were seven chromosomes which did not contain CAT interruptions. Two of these were from SCA1 subjects with pure CAG expansions (46 and 54 CAG repeat-units) and typical clinical features. The remaining five chromosomes were observed from control subjects without ataxia. The frequency of normal CAG repeat length with pure stretch was calculated as 1.3%. The relation between the length of CAG repeat configuration (i.e., combined CAG and CAT repeats) and presence/absence of CAT interruptions is summarized in Table 2 (Note: this table includes three atypical SCA1 subjects, as described later. Therefore, the total number of chromosomes becomes 390, the number of normal chromosomes is 385, and the number of SCA1 chromosomes is five). Of note is that we did not find any alleles with three CAT interruptions.
When the SfNaI digestion was performed in the dual fluorescence labeled PCR-restriction fragment length analysis, we were able to determine where CAT interruptions were present in the CAG repeat configuration (Fig. 3B). The CAG repeat length in the FITC-labeled 5′ fragment ranged in length from 10 up to 20 repeats in control chromosomes. This would indicate that the normal CAG repeat-unit lying in the 5′-region of the CAT interruption ranges between 10 and 20. Similarly, the CAG repeat length in the VIC-labeled 3′ fragment ranged in length from 7 to 23 repeats (Table 3).
Identification of a family with 40 CAG repeats with interruption
In our series of examining 193 individuals by the dual fluorescence labeled PCR-restriction fragment length analysis, we encountered an individual who harbored an allele with very small CAG repeat expansion. The size of this allele was 40 combined CAG and CAT repeats, which could be diagnosed as SCA1 by the classical criteria. By the dual fluorescent labeled PCR-restriction fragment length analysis, however, the repeat configuration was suggested to have two CAT interruptions with 27 CAG repeats in the 5′-end, and 10 CAG repeats in the 3′-end (“5′-CAG27CATCAGCATCAG10-3′”). When the PCR product was sub-cloned into PCR-TOPO and sequenced, the allele with 40 CAG repeat configuration was confirmed to harbor two CAT interruptions as expected from the dual fluorescent labeled PCR-restriction fragment length analysis. From the distribution pattern of actual CAG repeat number found in control Japanese (Fig. 3B), we considered that this particular individual had an abnormal SCA1 allele.
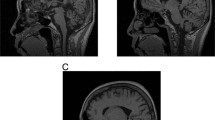
This patient was a 47-year-old male subject showing gait disturbance due to marked spastic paraparesis and mild truncal ataxia. He first noticed difficulties in walking at the age of 43. He had two elder siblings with similar neurological symptoms. The patient’s mother, who died at the age of 75, also showed progressive gait disturbance beginning from her fourth decade. Although a DNA sample of this mother was not available for examination, DNA samples of the patient’s siblings were tested and were both confirmed to have the same allele with “5′-CAG27CATCAGCATCAG10-3′”. Magnetic resonance imaging of the brain and the cervical, thoracic and lumbar spinal cord of these three subjects revealed mild cerebellar atrophy without obvious spinal cord or brainstem atrophy, which is compatible with SCA1 (Burk et al.1996). We also tested for mutations in spastin gene (Proukakis et al.2003), the most common gene identified for autosomal dominant spastic paraplegia (Svenson et al.2001). However, there was no mutation at least in the coding region of this gene (data shown upon request). From these observations, we conclude that spastic ataxia phenotype in these patients carrying 40 CAG repeat configuration (“5′-CAG27CATCAGCATCAG10-3′”) may be caused by a mild CAG repeat expansion in the ATXN1 gene.
Discussion
The main fruit of this study is the development of a new diagnostic method that would allow ones to detect actual CAG repeat numbers and the number of CAT interruptions in the CAG repeat configuration. The conventional method using a primer pair flanking CAG repeat configuration allowed one to measure total CAG and CAT repeat-units (Goldfarb et al. 1996; Jodice et al. 1997; Matsuyama et al. 1999; Pujana et al. 1999; Zhulke et al. 2002).
Sobczak and Krzyzosiak (2004) developed a new method, “SSCP-duplex analysis”, and showed exact CAG repeat configurations in 50 Polish individuals. Hellenbroich and his colleague developed a method that would detect actual CAG repeat by using non-labeled primers and SfaNI digestion (Zhulke et al. 2002). They studied in their German population and found rare alleles with mild CAG repeat expansion. However, their method would not discriminate 5′-end SfaNI-digested fragment with 3′-end SfaNI-digested fragment, and would need another step to disclose true CAG repeat configuration. In contrast, our method could allow one to discriminate both 5′- and 3′- fragments by labeling sense and anti-sense primers with different fluorescent dyes. Therefore, it would be much convenient and accurate to determine actual CAG repeat-unit encoding polyglutamine tract by employing our dual fluorescence labeled PCR-restriction fragment length analysis on fluorescence sequencers.
By using this new method, we have shown CAG repeat configurations in 385 control Japanese chromosomes and five SCA1 patients chromosomes. As has been reported in many studies, most of the normal chromosomes (98.7%) contain at least one CAT interruption in the CAG repeat configuration (Chung et al. 1993). However, we also found that 5 out of 385 (1.3%) control Japanese chromosomes do not contain the interruption. Since presence of CAT interruptions is considered to stabilize the CAG repeat length, presence of normal alleles without the CAT interruption may have some effect in the emergence of CAG repeat expansion through transmission. Comparing repeat numbers of CAG repeat configuration with and without CAT interruptions, alleles with interruption tended to have longer CAG repeat. In other words, it was not clear whether pure CAG repeat expansion occurs from the normal CAG repeat without CAT interruption.
In conclusion, the present method could be a convenient way to detect the accurate number of CAG repeat-unit encoding polyglutamine tract. The usefulness would be particularly important for CAG repeats with borderline length.
References
Banfi S, Servadio A, Chung MY, Kwiatkowski TJ Jr, McCall AE, Duvick LA, Shen Y, Roth EJ, Orr HT, Zoghbi HY (1994) Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet 7:513–520
Burk K, Abele M, Fetter M, Dichgan J, Skalej M, Laccone F, Didierjean O,Brice A, Klockgether T (1996) Autosomal dominant cerebellar ataxia type I clinical features and MRI in families with SCA1, SCA2 and SCA3. Brain 119(Pt 5):1497–1505
Chung M-Y, Ranum LPW, Duvick LA, Servadio A, Zoghbi HY, Orr HT (1993) Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type 1. Nat Genet 5:254–258
Quan F, Janas J, Poppvich BW (1995) A novel CAG repeat configuration in the SCA1 gene: implications for the molecular diagnostics of spinocerebellar ataxia type 1. Hum Mol 4:2411–2413
Goldfarb LG, Vasconcelos O, Platonov FA, Lunkes A, Kipnis V, Kononova S, Chabrashvili T, Vlasimirtsev VA, Alexeev VP, Gajdusek DC (1996) Unstable triplet repeat and phenotypic variability of spinocerebellar ataxia type 1. Ann Neurol 39:500–506
Ishikawa K, Tanaka H, Saito M, Ohkoshi N, Fujita T, Yoshizawa K, Ikeuchi T,Watanabe M, Hayashi A, Takiyama Y, Nishizawa M, Nakano I, Matsubayashi K, Miwa M, Shoji S, Knazawa I, Tsuji S, Mizusawa H (1997) Japanese families with autosomal dominant pure cerebellar ataxia map to chromosome 19p13.1–p13.2 and are strongly associated with mild CAG expansions in the spinocerebellar ataxia type 6 gene in chromosome 19p13.1. Am J Hum Genet 61(2):336–346
Jodice C, Giovannone B, Calabresi V, Bellocchi M, Terrenato L, Novelletto A (1997) Population variation analysis at nine loci containing expressed trinucleotide repeats. Ann Hum Genet 61:425–438
Li M, Ishikawa K, Toru S, Tomimitsu H, Takashima M, Goto J, Takiyama Y, Sasaki H, Imoto I, Inazawa J, Toda T, Kanazawa I, Mizusawa H (2003) Physical map and haplotype analysis of 16q-linked autosomal dominant cerebellar ataxia (ADCA) type III in Japan. Am J Hum Genet 48(3):111–118
Matilla T, Volpini V, Genis D, Rosell J, Corral J, Dávalos A, Molins A, Estivill X (1993) Presymptomatic analysis of spinocerebrllar ataxia type 1 (SCA1) via the expansion of the SCA1 CAG repeat in a large pedigree displaying anticipation and parental male bias. Hum Mol Genet 2:2123–2128
Matsuyama Z, Izumi Y, Kameyama M, Kawakami H, Nakamura S (1999) The effect of CAT trinucleotide interruptions on the age at onset of spinocerebellar ataxia type 1 (SCA1). J Med Genet 36:546–548
Orr H, Chung M-Y, Banfi S, Kwiatkowski TJ Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LPW, Zoghbi HY (1993) Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 4:221–226
Proukakis C, Auer-Grumbach M, Wagner K, Wilkinson PA, Reid E, Patton MA, Warner TT, Crosby AH (2003) Screening of patients with hereditary spastic paraplegia reveals seven novel mutations in the SPG4 (Spastin) gene. Hum Mutat 21(2):170
Pujana MA, Corral J, Gratacos M, Combarros O, Berciano J, Genis D, Banchs I, Estivill X, Volpini V (1999) Spinocerebellar ataxias in Spanish patients: genetic analysis of familial and sporadic cases. The ataxia study group. Hum Genet 104:516–522
Ranum LP, Chung M-Y, Banfi S, Bryer A, Schut LJ, Ramesar R, Duvick LA, McCall A, Subramony SH, Goldfarb L, Gomez C, Sandkuijl LA, Orr HT, Zoghbi HY (1994) Molecular and clinical correlations in spinocerebellar ataxia type 1: evidence for familial effects on the age at onset. Am J Hum Genet 55:244–252
Sobczak K, Krzyzosiak WJ (2004) Pattern of CAG repeat interruptions in SCA1 and SCA2 genes in relation to repeat instability. Hum Mutat 24:236–247
Svenson IK, Ashley-Koch AE, Gaskell PC, Riney TJ, Cumming WJ, Kingston HM, Hogan EL, Boustany RM, Vance JM, Nance MA, Pericak-Vance MA,Marchuk DA (2001) Identification and exoression analysis of spastin gene mutation in hereditary spastic paraplegia. Am J Hum Genet 68(5):1077–85
Tsuda H, Jafar-Nejad H, Patel AJ, Sun Y, Chen HK, Rose MF, Venken KJ, Botas J, Orr HT, Bellen HJ, Zoghbi HY (2005) The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gif-1/senseless proteins. Cell 122:633–644
Zhulke C, Dalski A, Hellenbroich Y, Bubel S, Schwinger E, Burk K (2002) Spinocerebellar ataxia type 1 (SCA1): phenotype-genotype correlation studies in intermediate alleles. Eur J Hum Genet 10:204–209
Author information
Authors and Affiliations
Corresponding author
Additional information
Analysis of CAG repeats in the ataxin-1 gene.
Rights and permissions
About this article
Cite this article
Lin, J.X., Ishikawa, K., Sakamoto, M. et al. Direct and accurate measurement of CAG repeat configuration in the ataxin-1 (ATXN-1) gene by “dual-fluorescence labeled PCR-restriction fragment length analysis”. J Hum Genet 53, 287–295 (2008). https://doi.org/10.1007/s10038-007-0242-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-007-0242-2
Keywords
This article is cited by
-
STRs: Ancient Architectures of the Genome beyond the Sequence
Journal of Molecular Neuroscience (2021)
-
Brain pathology of spinocerebellar ataxias
Acta Neuropathologica (2012)
-
Genetic and clinical analysis in a Chinese parkinsonism-predominant spinocerebellar ataxia type 2 family
Journal of Human Genetics (2011)