Abstract
Methotrexate is administered in high doses to treat childhood acute lymphoblastic leukemia and malignant lymphoma. Hepatotoxicity and bone marrow suppression often limit its use, however. The objective of this study was to determine the genetic polymorphisms associated with the hepatotoxicity and elimination of methotrexate. Genetic polymorphisms of glutathione S-transferase (GST) genes including GSTT1 positive/null, GSTM1 positive/null, and GSTP1 A313G, and genes for reduced folate carrier 1 G80A (RFC1 G80A), methylenetetrahydrofolate reductase C677T (MTHFR C677T), and breast cancer resistant protein C421A (BCRP C421A) were determined for 26 patients by the polymerase chain reaction (PCR) method or by direct sequencing. A high frequency of hepatotoxicity (P = 0.035) was observed for patients with GSTM1 positive and RFC1 AA80, and serum concentrations of methotrexate 48 h after the start of infusion were higher for patients with the TT677 genotype of MTHFR (P = 0.028). In conclusion, GSTM1 positive/null and RFC1 G80A polymorphisms could be predictors for hepatotoxicity, and the MTHFR C677T polymorphism is associated with elimination of methotrexate.
Similar content being viewed by others
Introduction
Childhood acute lymphoblastic leukemia (ALL) is characterized by uncontrolled proliferation and maturation arrest of lymphoid progenitor cells in the bone marrow. Marked advances in chemotherapy for childhood ALL have resulted in an expected cure rate of 80% (Pui et al. 2003; Aplenc and Lange 2004). Malignant lymphoma (ML) is also treated by chemotherapy with a high cure rate, although outcome is believed to depend on stage, histological type, etc. (Murphy et al. 1989; Anderson et al. 1993).
High-dose treatment with methotrexate often causes hepatotoxicity and bone marrow suppression, however (Schmiegelow et al. 1989; Schmiegelow and Pulczynska 1990), necessitating dose reduction or cessation of treatment, one reason for a relapse of ALL and ML. For appropriate use of methotrexate it would therefore be useful to identify a predictor and/or the mechanism of the adverse effects of methotrexate. In this work we focused on genetic polymorphisms of glutathione S-transferases (GSTs) including GSTT1 positive/null, GSTM1 positive/null, and GSTP1 A313G, reduced folate carrier 1 G80A (RFC1 G80A), methylenetetrahydrofolate reductase C677T (MTHFR C677T), and breast cancer resistant protein C421A (BCRP C421A).
Information on the genetic polymorphisms associated with the toxicity and efficacy of drugs has recently been accumulating (Donnelly 2004). It has been reported that the idiosyncratic severe hepatotoxicity of troglitazone, which was used to treat type-2 diabetes mellitus, is associated with genetic polymorphisms of GSTM1 and GSTT1 (Watanabe et al. 2003). Patients without GSTM1 and GSTT1 were more at risk of hepatotoxicity of troglitazone. GSTs are thought to detoxify by conjugation of xenobiotics with glutathione. Patients with the null type of GSTM1 were at increased risk of heptatotoxicity of antituberculosis drugs (Roy et al. 2001). Null types of GSTT1 and GSTM1 have also been associated with an increase in the frequency of hepatotoxicity of tacrine, an anticholinesterase inhibitor (Simon et al. 2000). In contrast, lack of GSTP1 has been related to the toxicity of acetaminophen in mice (Henderson et al. 2000). The genetic polymorphism at A313G of GSTP1, resulting in amino acid substitution, has been reported to be associated with enzymatic activity (Watson et al. 1998). It is generally believed that polymorphisms of GSTs could be factors affecting drug-induced toxicity.
Reduced folate carrier 1, the function of which is to transport reduced folate, is expressed ubiquitously throughout the body. Methotrexate is a substrate for RFC1 and its distribution to tissue is thought to be mediated by RFC1. RFC1 expression levels (Levy et al. 2003) and genetic polymorphisms of RFC1 G80A (Laverdiere et al. 2002) have been reported to be associated with ALL outcome, and the latter has also been associated with plasma methotrexate levels in patients with ALL. Here we have evaluated the association of the polymorphism G80A in RFC1 with hepatotoxicity after high-dose treatment with methotrexate.
MTHFR is a key enzyme in the metabolism of folate. It has been reported that genetic polymorphism at C677T of MTHFR is associated with the toxicity of methotrexate in patients with bone marrow transplants (Ulrich et al. 2001), rheumatoid arthritis (Urano et al. 2002), and ovarian cancer (Toffoli et al. 2003).
Breast cancer resistant protein (BCRP, ABCG2) is an ABC transporter with an ATP binding cassette. It has been reported to remove substrates from cells to confer resistance to anticancer agents including methotrexate (Doyle and Ross 2003; Abbott 2003). It is also believed that BCRP in the bile canalicular membrane of the liver is a key determinant of the disposition to secrete substrates of a drug into bile. Genetic polymorphism at C431A of BCRP has been reported to be associated with expression and function in vitro and in vivo (Imai et al. 2002; Mizuarai et al. 2004; Kobayashi et al. 2005).
Here we have investigated the effects of genetic polymorphisms of positive/null GSTT1 and positive/null GSTM1, and GSTP1 A313G, RFC1 G80A, MTHFR C677T, and BCRP C421A on the hepatotoxicity and elimination of methotrexate in patients with childhood ALL or ML after high-dose treatment with the drug.
Materials and methods
Subjects
Blood samples were obtained from 26 patients with ALL (n = 20) or ML (n = 6) who were treated at Kobe University Hospital, Japan (n = 26, 16 males and 10 females). The patients were administered a high dose of methotrexate (3 g m−2) followed by leucovorin rescue treatment in the event of complete remission. If the methotrexate concentration 48 h after starting the administration was higher than 1 μmol L−1, additional leucovorin rescue was performed. The age range was 10 months to 15 years, average 6.7 ± 4.7. Informed consent was obtained from all the patients or their guardians before the study. The study procedure was approved by the institutional review board of Kobe University Hospital, Kobe University, Japan.
Genotyping for GSTT1, GSTM1, GSTP1, RFC1, MTHFR, and BCRP
Genomic DNA was extracted from blood samples by use of a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and from buccal mucosa using a Smitest EX R&D DNA extraction kit (Genome Science Laboratories, Fukushima, Japan), according to the manufacturer’s directions. Table 1 lists the primer pairs used in this study. Each PCR was performed with 30 ng DNA, ExTaq Buffer, 0.5 mmol L−1 dNTP mixture, 1.25 U ExTaq polymerase (Takara Bio, Shiga, Japan), and 0.4 μmol L−1 each of the forward and reverse primers in a 25 μL reaction volume. PCR cycling was programmed as shown in Table 1. For GSTT1 and GSTM1, PCR products were electrophoresed in 3% agarose gel (Takara Bio) and positive/null genotypes were determined by the position of bands. For determination of genotypes of GSTP1, RFC1, MTHFR, and BCRP, PCR products were used for direct sequencing. They were treated with Applied Biosystems Version 1.1 BigDye terminator chemistry. The reaction mixtures were purified with a CentriSep column (Promceton Separations, Adelphia, NJ, USA). The products were electrophoresed for sequencing on an Applied Biosystems 3100 Avant Genetic Analyzer (Applied Biosystems). All sequence chromatograms were read in both the sense and antisense directions.
Assessment of hepatotoxicity
Laboratory test values were monitored routinely and alanine aminotransferase (ALT) activity was used to assess hepatotoxicity. Toxicity was graded on the basis of National Cancer Institute Common Toxicity Criteria (Ver 2.0) using maximum ALT levels within one week of administration of methotrexate. Relationships between genetic polymorphisms and hepatotoxicity of methotrexate were investigated using the data for 18 patients whose ALT levels were within the normal range (≥34) before methotrexate dosing.
Serum concentration of methotrexate
Serum concentrations of methotrexate were measured with a TDx analyzer (Abbott Japan, Tokyo, Japan).
Statistical analysis
Statistical analysis of the association of genetic polymorphisms with hepatotoxicity or serum concentrations were performed with Fisher’s extended exact test.
Results
HD-MTX resulted in grade 2/3 hepatotoxicity in five of eighteen patients, three of eleven for males and two of seven for females. The mean ages of patients without and with MTX-induced hepatotoxicity were 5.5 ± 4.5 and 8.0 ± 4.8, respectively. These results suggest that gender and age affect susceptibility to MTX-induced hepatotoxicity, although larger studies are needed to confirm this.
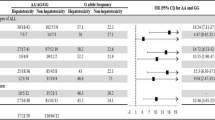
Table 2 shows frequencies for the genotypes of GSTM1, GSTT1, GSTP1, RFC1, MTHFR, and BCRP. Table 3 shows the association of genetic polymorphisms of these genes with hepatotoxicity. No significant association was found on the basis of the genotypes of one gene. Table 4 shows the effect of RFC1 genotypes on hepatotoxicity in patients carrying GSTM1, indicating that patients with AA80 are likely to suffer from hepatotoxicity (P < 0.05; Fisher’s extended exact test).
To investigate the genetic factors affecting elimination of methotrexate, association of genetic polymorphisms with the serum concentration of methotrexate 48 h after intravenous infusion at a dose of 3 g m−2 was evaluated. Although there was no association for GSTM1, GSTT1, GSTP1, RFC1, or BCRP, significantly more of patients whose concentration remained >1 μmol L−1 at 48 h had the TT677 genotype (P < 0.05; Fisher’s exact test) (Table 5).
Discussion
Acute lymphoblastic leukemia is the most common malignancy in children. Recent advances in chemotherapy have resulted in a cure rate of approximately 80% (Pui et al. 2003; Aplenc and Lange 2004). ML is also treated by chemotherapy, with a high cure rate (Murphy et al. 1989; Anderson et al. 1993). Treatment is, however, often limited by severe toxicity, including hepatotoxicity and myeloid suppression (Schmiegelow et al. 1989; Schmiegelow and Pulczynska 1990). It is therefore important to predict the occurrence of adverse events. Here, we investigated genetic polymorphisms to predict the hepatotoxicity of methotrexate.
We showed that the genetic polymorphism G80A in RFC1 was associated with hepatotoxicity in patients carrying GSTM1. These results suggested that RFC1 is a key enzyme determining the residual amount of methotrexate in the liver. It is thought that the residual amount of methotrexate should be higher in patients with an A allele at position 80 in RFC1, which might reduce the activity of RFC1, resulting in a greater risk of methotrexate-induced hepatotoxicity. GSTs are believed to be responsible for reducing the toxicity of xenobiotics. Patients with null types of GSTM1 and/or GSTT1 are reportedly at higher risk of hepatotoxicity of troglitazone (Watanabe et al. 2003), antituberculosis drugs (Roy et al. 2001), and tacrine, an anticholinesterase inhibitor (Simon et al. 2000), suggesting that a lack of or reduction of GSTs increases the risk of drug induced-hepatotoxicity. Our current results also suggest that carrying GSTM1 results in a greater risk of methotrexate-induced hepatotoxicity. No reasonable explanation of this is yet available, but GSTP has been reported to increase the risk of hepatotoxicity from acetaminophen by inhibition of Jun N-terminal kinase (Adler et al. 1999). In our study hepatotoxicity tended to be more frequent in patients with the T allele at 677, although this was not statistically significant. Toffoli et al. (2003) reported association of MTHFR C677T with severe toxicity of MTX, including neutropenia, anemia, and mucositis in ovarian cancer patients, but they did not report data for hepatotoxicity. Chiusolo et al. (2002) have also shown that the MTHFR C677T genotype is associated with hematopoietic and hepatic toxicity. On the basis of their results and ours, the MTHFR genotype might lead to susceptibility to MTX hepatotoxicity, although more studies should be conducted.
We also found a significant association of C677T in MTHFR with the residual concentration of methotrexate at 48 h. Because elimination of methotrexate at the terminal phase is thought to be associated with the toxicity of methotrexate (Stoller et al. 1977), additional leucovorin rescue is usually performed when the concentration of methotrexate was higher than 1 μmol L−1. Our current findings suggest that genotyping of MTHFR could enable us to predict the need for additional leucovorin rescue, although further clinical study is necessary. MTHFR is not thought to be responsible for the metabolism or transport of methotrexate. The mechanism has not been clarified, but Toffoli et al. (2003) showed that plasma concentrations of homocysteine after methotrexate dosing were higher in patients with the TT genotype at MTHFR C677T. Homosysteine concentration might be useful for discussion of the association of this genotype with MTX concentration.
Laverdiėre et al. (2002) reported higher concentrations of MTX in patients with AA at G80A of RFC1, leading poor prognosis. We did not find association of this genotype with MTX concentration. The reason for the discrepancy is unclear, although it is possibly because of sample size.
In conclusion, we have shown that genetic polymorphisms of RFC1 G80A and GSTM1 positive/null are risk factors for methotrexate hepatotoxicity and polymorphism of MTHFR C677T is associated with the serum concentration of methotrexate. These results are useful for treatment of ALL by use of methotrexate.
References
Abbott BL (2003) ABCG2 (BCRP) expression in normal and malignant hematopoietic cells. Hematol Oncol 21:115–130
Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, Pincus MR, Sardana M, Henderson CJ, Wolf CR, Davis RJ, Ronai Z (1999) Regulation of JNK signaling by GSTp. EMBO J 18:1321–1334
Anderson JR, Jenkin RD, Wilson JF, Kjeldsberg CR, Sposto R, Chilcote RR, Coccia PF, Exelby PR, Siegel S, Meadows AT (1993) Long-term follow-up of patients treated with COMP or LSA2L2 therapy for childhood non-Hodgkin’s lymphoma: a report of CCG-551 from the Children’s Cancer Group. J Clin Oncol 11:1024–1032
Aplenc R, Lange B (2004) Pharmacogenetic determinants of outcome in acute lymphoblastic leukaemia. Br J Haematol 125:421–434
Chiusolo P, Reddiconto G, Casorelli I, Laurenti L, Sora F, Mele L, Annino L, Leone G, Sica S (2002) Preponderance of methylenetetrahydrofolate reductase C677T homozygosity among leukemia patients intolerant to methotrexate. Ann Oncol 13:1915–1918
Donnely JG (2004) Pharmacogenetics in cancer chemotherapy. Ther Drug Monit 26:231–235
Doyle LA, Ross DD (2003) Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2). Oncogene 22:7340–7358
Henderson CJ, Wolf CR, Kitteringham N, Powell H, Otto D, Park BK (2000) Increased resistance to acetaminophen hepatotoxicity in mice lacking glutathione S-transferase Pi. Proc Natl Acad Sci USA 97:12741–12745
Imai Y, Nakane M, Kage K, Tsukahara S, Ishikawa E, Tsuruo T, Miki Y, Sugimoto Y (2002) C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol Cancer Ther 1:611–616
Kobayashi D, Ieiri I, Hirota T, Takane H, Maegawa S, Kigawa J, Suzuki H, Nanba E, Oshimura M, Terakawa N, Otsubo K, Mine K, Sugiyama Y (2005) Functional assessment of ABCG2 (BCRP) gene polymorphisms to protein expression in human placenta. Drug Metab Dispos 33:94–101
Laverdiėre C, Chiasson S, Costea I, Moghrabi A, Krajinovic M (2002) Polymorphism G80A in the reduced folate carrier gene and its relationship to methotrexate plasma levels and outcome of childhood acute lymphoblastic leukemia. Blood 100:3832–3834
Levy AS, Sather HN, Steinherz PG, Sowers R, La M, Moscow JA, Gaynon PS, Uckun FM, Bertino JR, Gorlick R, Children’s Cancer Group Study Children’s Cancer Group Study (2003) Reduced folate carrier and dihydrofolate reductase expression in acute lymphocytic leukemia may predict outcome: a children’s cancer group study. J Pediatr Hematol Oncol 25:688–695
Mizuarai S, Aozasa N, Kotani H (2004) Single nucleotide polymorphisms result in impaired membrane localization and reduced ATPase activity in multidrug transporter ABCG2. Int J Cancer 109:238–246
Murphy SB, Fairclough DL, Hutchison RE, Berard CW (1989) Non-Hodgkin’s lymphomas of childhood: an analysis of the histology, staging, and response to treatment of 338 cases at a single institution. J Clin Oncol 7:186–193
Pui CH, Sandlund JT, Pei D, Rivera GK, Howard SC, Ribeiro RC, Rubnitz JE, Razzouk BI, Hudson MM, Cheng C, Raimondi SC, Behm FG, Downing JR, Relling MV, Evans WE (2003) Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA 290:2001–2007
Roy B, Chowdhury A, Kundu S, Santra A, Dey B, Chakraborty M, Majumder PP (2001) Increased risk of antituberculosis drug-induced hepatotoxicity in individuals with glutathione S-transferase M1 ‘null’ mutation. J Gastroenterol Hepatol 16:1033–1037
Schmiegelow K, Pulczynska M (1990) Prognostic significance of hepatotoxicity during maintenance chemotherapy for childhood acute lymphoblastic leukaemia. Br J Cancer 61:767–772
Schmiegelow K, Schroder H, Pulczynska MK, Hejl M (1989) Maintenance chemotherapy for childhood acute lymphoblastic leukemia: relation of bone-marrow and hepatotoxicity to the concentration of methotrexate in erythrocytes. Cancer Chemother Pharmacol 25:65–69
Simon T, Becquemont L, Mary-Krause M, de Waziers I, Beaune P, Funck-Brentano C, Jaillon P (2000) Combined glutathione-S-transferase M1 and T1 genetic polymorphism and tacrine hepatotoxicity. Clin Pharmacol Ther 67:432–437
Stoller RG, Hande KR, Jacobs SA, Rosenberg SA, Chabner BA (1977) Use of plasma pharmacokinetics to predict and prevent methotrexate toxicity. N Engl J Med 297:630–634
Toffoli G, Russo A, Innocenti F, Corona G, Tumolo S, Sartor F, Mini E, Boiocchi M (2003) Effect of methylenetetrahydrofolate reductase 677C → T polymorphism on toxicity and homocysteine plasma level after chronic methotrexate treatment of ovarian cancer patients. Int J Cancer 103:294–299
Ulrich CM, Yasui Y, Storb R, Schubert MM, Wagner JL, Bigler J, Ariail KS, Keener CL, Li S, Liu H, Farin FM, Potter JD (2001) Pharmacogenetics of methotrexate: toxicity among marrow transplantation patients varies with the methylenetetrahydrofolate reductase C677T polymorphism. Blood 98:231–234
Urano W, Taniguchi A, Yamanaka H, Tanaka E, Nakajima H, Matsuda Y, Akama H, Kitamura Y, Kamatani N (2002) Polymorphisms in the methylenetetrahydrofolate reductase gene were associated with both the efficacy and the toxicity of methotrexate used for the treatment of rheumatoid arthritis, as evidenced by single locus and haplotype analyses. Pharmacogenetics 12:183–190
Watanabe I, Tomita A, Shimizu M, Sugawara M, Yasumo H, Koishi R, Takahashi T, Miyoshi K, Nakamura K, Izumi T, Matsushita Y, Furukawa H, Haruyama H, Koga T (2003) A study to survey susceptible genetic factors responsible for troglitazone-associated hepatotoxicity in Japanese patients with type 2 diabetes mellitus. Clin Pharmacol Ther 73:435–455
Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA (1998) Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 19:275–280
Acknowledgments
This study was supported in part by a Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Imanishi, H., Okamura, N., Yagi, M. et al. Genetic polymorphisms associated with adverse events and elimination of methotrexate in childhood acute lymphoblastic leukemia and malignant lymphoma. J Hum Genet 52, 166–171 (2007). https://doi.org/10.1007/s10038-006-0096-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-006-0096-z
Keywords
This article is cited by
-
Effects of gene polymorphisms on delayed MTX clearance, toxicity, and metabolomic changes after HD-MTX treatment in children with acute lymphoblastic leukemia
European Journal of Pediatrics (2023)
-
Population Pharmacokinetic Model of Methotrexate in Brazilian Pediatric Patients with Acute Lymphoblastic Leukemia
Pharmaceutical Research (2023)
-
Effects of splicing-regulatory polymorphisms in ABCC2, ABCG2, and ABCB1 on methotrexate exposure in Chinese children with acute lymphoblastic leukemia
Cancer Chemotherapy and Pharmacology (2023)
-
A Systematic Review of Population Pharmacokinetic Models of Methotrexate
European Journal of Drug Metabolism and Pharmacokinetics (2022)
-
Associations between the C677T and A1298C polymorphisms of MTHFR and the toxicity of methotrexate in childhood malignancies: a meta-analysis
The Pharmacogenomics Journal (2018)