Abstract
β-thalassemia is a common monogenic disease caused by mutations in the human β-globin gene (HBB), many of which are differentially represented in human subpopulations stratified by ethnicity. This study describes an efficient and highly accurate method to screen for the eight most-common disease-causing mutations, covering more than 98% of HBB alleles in the Taiwanese population, using parallel minisequencing and multiplex assay by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The MALDI-TOF MS was optimized for sensitivity and resolution by “mass tuning” the PinPoint assay for eight HBB SNPs. Because of the close proximity and clustering of mutations in HBB, primer extension reactions were conducted in parallel. Efficient sequential desalting using POROS and cationic exchange chromatography allowed for an unambiguous multiplex genotyping by MALDI-TOF MS. The embellishing SNP assay allowed for highly accurate identification of the eight most-common β-thalassemia mutations in homozygous normal control, carrier, and eight heterozygous carrier mixtures, as well as the diagnosis of a high-risk family. The results demonstrated a flexible strategy for rapid identification of clustering SNPs in HBB with a high degree of accuracy and specificity. It can be adapted easily for high-throughput diagnosis of various hereditary diseases or to establish family heritage databases for clinical applications.
Similar content being viewed by others
Introduction
β-thalassemia, a common hereditary disease found throughout the world, is caused by the mutation of a single gene (Weatherall and Clegg 1996). Although large deletions are sometimes observed, most β-thalassemia-causing mutations involve one or several nucleotides of the β-globin gene (HBB; MIM # 141900) (Henthorn et al. 1990; Kazazian 1990). Currently, approximately 200 different mutations have been reported worldwide to be associated with this disease, and various ethnic groups affected by β-thalassemia each display their own particular common mutation(s) (Abdullah et al. 1996; Baysal and Carver 1995; Ko et al. 1997) as well as a variable number of rare alleles (Weatherall and Clegg 2001). For example, in the Southeast Asian (Weatherall and Clegg 2001) and Chinese populations, five β-globin gene alleles (CD41/42 –TCTT, IVS-2 +654 C>T, 28 A>G, CD17 A>T, and CD71/72 +A) account for more than 90% of all β-thalassemia-causing mutations (Ko and Xu 1998). Together with the three other β-globin gene alleles (CD27/28 +C, CD43 G>T, and CD1 T>G), these eight SNPs account for more than 98% of HBB mutations in the Taiwanese population (Su et al. 2003). Given that β-thalassemia is caused by known, specific mutations of the HBB gene, the development of a highly accurate, rapid, high-throughput, and direct assay for the simultaneous genotyping of multiple common HBB alleles associated with β-thalassemia is important for the diagnosis of this worldwide disease.
Several assays have been developed to genotype common β-thalassemia mutations, including amplification refractory mutation analysis (Fortina et al. 1992), DNA-probe assays (Ugozzoli et al. 1998), fluorescent multiplex PCR (Sherlock et al. 1998), real-time PCR (Vrettou et al. 2004), and microelectronic chip-based assays (Foglieni et al. 2004). Each of these methods has advantages and limitations, and a highly accurate, high-throughput method to simultaneously genotype common HBB alleles has yet to be developed and widely implemented.
Minisequencing is a direct method that can be used to generate allele-specific DNA products, which is potentially useful for multiplex high-throughput genotyping (Syvanen 1999; Tan et al. 2001). Relatively short primer extension products templated on experimental samples can be produced and analyzed rapidly. Control templates with the wild-type or mutant target sequence can be used to generate markers that identify each genotype of interest. Minisequencing has been widely applied for multiplex genotyping of common β-thalassemia mutations. These assays utilize several different methods to characterize the primer extension products, including denaturing HPLC (Su et al. 2003; Wu et al. 2003; Yip et al. 2003), solid-phase array (Kurg et al. 2000), or fluorescence-based gel capillary electrophoresis (Wang et al. 2003).
The introduction of matrix-assisted laser desorption/ionization (MALDI) time-of-fight (TOF) mass spectrometry (MS) has resulted in the direct and rapid identification of intact biomolecules (Marvin et al. 2003). For the analysis of complex minisequencing products in multiplex genotyping, MS offers greater accuracy than electrophoresis-based or hybridization-array-based methods, which are often susceptible to complications from nucleic acid secondary structure (Li et al. 1993; Southern et al. 1994). Therefore, MALDI-TOF MS in combination with a minisequencing strategy has become one of the most promising mutation analysis tools to analyze polymorphisms in disease-causing genes, including the genes encoding cystic fibrosis (Braun et al. 1997), coronary artery diseases (Nakai et al. 2002), cardiovascular disease, and even the disease-causing mutations on human Y chromosome (Paracchini et al. 2002; Wise et al. 2003). Compared with other DNA analysis methods, the advantages of MALDI-TOF MS include the direct and absolute mass readout of DNA (Tost and Gut 2002) and high-throughput analysis (Van Ausdall and Marshall 1998) that facilitates simultaneous multiplex genotyping in a single experiment.
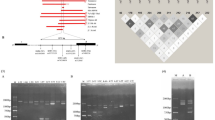
Although multiplex assay is generally a cost-effective way for high-throughput SNP genotyping, two factors have limited the practicality of using multiplex minisequencing and MALDI-TOF MS for screening HBB mutations. The limitations are based on the following factors: (1) difficult and time-consuming optimization of multiplex primer extension (Tost and Gut 2002), and (2) spectral interference from contaminating salt adducts (Guo 1999; Tost and Gut 2002). The former issue presents a specific challenge because highly multiplex genotyping parameters may be difficult to optimize. For HBB, as shown in Fig.1, two sets of clustering polymorphic sites, CD27/28 +C versus CD17 A>T, and CD43 G>T versus CD41/42 –TCTT, are in close proximity to each other and almost overlap. The genotyping of these closely associated and clustered mutations within the β-thalassemia gene poses practical difficulty for multiplex minisequencing in a single tube (Wang et al. 2003). The second limiting factor for multiplex HBB genotyping using MALDI-TOF MS is that contaminating salt, detergent, or glycerol can degrade spectral quality and reduce the accuracy of mass assignment (Bleicher and Bayer 1994; Guo 1999; Kim et al. 2002; Tost and Gut 2002). In a highly multiplex assay, this increase in interference is greater than the linear scale of the multiplex factor. Thus, sample purification is a crucial step in DNA sample analysis by MALDI-TOF MS.
Human β-globin gene sequence and primer locations. The mutation sites are marked in capital letters and red font. The shaded DNA sequences indicate primers. Assignments are shown in the right column and shaded in the same color as the relevant primer. Overlapping sequences are shaded in pink. The sequences for PCR amplification are shaded in cyan
To address the complications of primer extension arising from the close proximity and clustering of β-thalassemia mutations, we developed an alternative multiplex genotyping method utilizing eight parallel minisequencing reactions followed by multiplex MALDI-TOF analysis. Minsequencing reaction products are pooled for sequential desalting and MALDI-TOF MS assay, thus maintaining the advantages and efficiency of multiplex processing and analysis. Using this approach, the eight common HBB alleles in the Taiwanese population can be analyzed with high accuracy in a single assay. The method is equally successful with heterozygous carriers or homozygous-affected and normal individuals. Our results demonstrate the feasibility of rapid, accurate, and flexible multiplex genotyping for high throughput, unconstrained diagnostics of β-thalassemia.
Materials and methods
DNA sample preparation
Cases and controls are from the individuals who came to the National Taiwan University Hospital to be tested for the national screening program for thalassemia. The donors of all samples gave informed consent. All samples were extracted from peripheral whole blood, chorionic villus sampling, amniocentesis, or cord-blood sampling by using Puregene DNA Isolation Kit (Gentra Systems Inc., Minneapolis, MN, USA) according to the manufacturer’s instructions. The DNA samples represent common genotypes of the β-globin gene in the Taiwanese population. The diagnosis of β-thalassemia was confirmed previously by PCR-based techniques, including analyses of amplification-created restriction sites (ACRS) and sequence analysis.
Primers design and PCR condition
The PCR primers used in this study were: 5′-TACGGCTGTCATCACTTAGACCTCA-3′ and 5′-TGCAGCTTGTCACAGTGCAGCTCACT-3′ for amplification of exons 1 to 5′ portion of exons 2 and 5′-GTGTACACATATTGACCAAA-3′ and 5′-AGCACACAGACCAGCACGT-3′ for amplification of exon 3 and its flanking regions. According to previous reports (Ko et al. 1997), we were able to detect most frequently occurring mutations, apart from the deletion type, up to a level of 98% for the HBB mutations among the general Taiwanese population.
Genomic DNA (100 ng) template was amplified in a multiblock system (MBS) thermocycler (ThermoHybaid, Ashford, UK) in a total volume of 25 μl containing 0.12 μM of each primer, 100 μM of each dNTPs, 10 mM Tris–HCl (pH=8.3), 50 mM KCl, 2 mM MgCl2, and 0.5 units of AmpliTaq Gold enzyme (PE Applied Biosystems, Foster City, CA, USA). The initial denaturation step was set at 95°C for 10 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 1 min, extension at 72°C for 30 s, and final extension at 72°C for 10 min.
Preparation of PCR products for primer extension (minisequencing) reaction
The unincorporated dNTPs and primers from PCR products were removed by GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ, USA), then the purified PCR products was subsequently used for the primer extension reactions. The minisequencing extension primers for PinPoint assay, all four unlabeled ddNTPs, were included (Fei et al. 1998; Haff and Smirnov 1997a; Li et al. 1999) to detect the eight most-common genotypes of β-globin gene mutation, including CD41/42 –TCTT, IVS2 +654 C>T, −28 A>G, CD17 A>T, CD71/72 + A, CD27/28 +C, CD43 G>T, and CD1 T>G, are summarized in Table 1. Primer extension reactions were carried out in 20 μl solution containing 50 ng PCR product and 1 μl of 10 pmol minisequencing primer, 0.5 μl of 1 mM of each ddNTPs (Amersham Pharmacia Biotech, Piscataway, NJ, USA), 0.5 U of Thermo Sequenase DNA polymerase (Amersham Biosciences), and 2 μl of 10× reaction buffer provided by the manufacturer. The reaction was carried out in a multiblock system (MBS) thermocycler (ThermoHybaid) with an initial denaturation step at 96°C for 1 min followed by 50 cycles of 96°C for 15 s, 43°C for 15 s, 60°C for 100 s, then 96°C for 30 s.
Sample purification
Three different DNA desalting methods were tested for 20 μl of 10 μM single-stranded DNA primer (5′-AACTCATCCACGTTCACCT-3′) in 5 mM MgCl2. For ethanol precipitation (Sun et al. 2000), 3 M ammonium acetate solution of one-fifth to one-third sample volume was added to DNA and incubated in 65°C for 60 min and then 37°C for 2 h. After incubation, 10 volumes of 100% ethanol were added, incubated in a freezer at −20°C overnight followed by removing the supernatant solution with centrifugation. Finally, 5 μl of water was added to redissolve the precipitated DNA. For cationic exchange resin desalting, the 5WX8-200 Dowex cation exchange resin (Supelco Park, Bellefonte, PA, USA) was selected according to published protocol of Harksen et al. (1999). The cationic exchange beads were first exchanged with 1 M NH4OAc and placed on a piece of parafilm, and 5–10 μl of DNA sample and an equal amount of matrix (50 mg/ml 3-hydroxypicolinic acid, 3HPA) were added to 0.1 mg beads followed by mixing up and down in the pipette tip several times. The supernatant was loaded onto the sample plate for MALDI analysis. The third method for DNA purification uses POROS 50-R2 (PerSeptive Biosystems, Framingham, MA, USA) reversed-phase chromatography media (Haff and Smirnov 1997b). Pipette tips were filled with 20 μl POROS (80 mg/ml in 30% ethanol) and then replaced by 20 μl of 100 mM triethylammonium acetate (TEAA, pH=6.5). DNA sample was extracted in the presence of 100 mM TEAA (pH=6.5), rinsed with 100 mM and 10 mM TEAA (pH=6.5), and eluted with 20% acetonitrile. Solvent was removed by applying compressed air to the tip in all steps (Guo 1999).
For sequential desalting combining the POROS chromatography and cationic exchange resin, vacuum-dried sample following POROS purification protocol was resolved in 3–5 μl water and desalted following the cationic exchange bead procedure described above. For multiplex genotyping, 40 μl of each minisequencing product were pooled for sequential desalting procedure prior to spotting onto the MALDI sample plate.
Mass spectrometry analysis
Prior to MALDI-TOF MS analysis, the sample was mixed with matrix solution (50 mg/ml 3-hydropicolinic acid in a 4:5:1 mixture of water, acetonitrile and 50 mg/ml diammonium citrate) and spotted on the 96×2-well teflon sample plate (PerSeptive Biosystems). MALDI-TOF mass spectra were acquired by a reflectron TOF MS (Voyager-DE PRO, Applied Biosystems). The instrument was equipped with a 337 nm nitrogen laser source at 3–20 Hz. Measurements were taken in linear, negative ion mode at 20 kV acceleration voltage and 380 ns delayed ion extraction. The excess unextended oligonucleotide primers were used as internal standards for mass calibration. A typical mass spectrum was obtained by the average of 100 laser shots followed by noise reduction and Gaussian smoothing using Data Explorer software (Applied Biosystems).
Sequence analysis
Amplicons were purified by solid-phase extraction and bidirectionally sequenced with the PE Biosystems Taq DyeDeoxy terminator cycle sequencing kit (PE Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Sequencing reactions were separated on a PE Biosystems 373A/3100 sequencer.
Results
Rational design for HBB multiplex genotyping primers
Multiplex genotyping is an important strategy to reduce cost and improve efficiency in screening for common disease-causing mutations in HBB. This approach is used here to identify homozygotes and heterozygotes (normal and affected individuals) carrying the eight common HBB alleles in the Taiwanese population. PinPoint assay was chosen as the primer extension reaction where the primer is extended by a single nucleotide in the presence of all four dideoxynucleotides (Fei et al. 1998; Haff and Smirnov 1997a; Li et al. 1999). In this assay, the primers and their extension products are in a narrow mass range, which facilitates the display and interpretation of eight SNPs in one mass spectrum. To resolve the eight pairs of primer/extended products without ambiguity, the overlap of different pairs of unextended primer and product must be avoided. This can be achieved by the rational design of the primers using a “mass tuning” strategy in which the primers vary with successively increasing length. For our study, primer lengths were designed with at least one base difference, ranging from 18 to 27 bases. Table 1 summarizes the masses of the designed mutation-specific primers and the corresponding extension products.
Optimization of desalting prior to MALDI-TOF MS
Because of the small mass difference at the polymorphic site using the PinPoint assay, the spectrum may be difficult to interpret if salt adducts accompany targeting peaks. Additionally, the ion suppression caused by excess salt will reduce the sensitivity of mass spectrometric measurement. Therefore, efficient desalting of the samples will be crucial for multiplex genotyping to remove the salts and buffer used in the primer extension reactions.
To address the issue, a variety of sample purification methods, including ethanol precipitation, cationic exchange resin, and reverse-phase resin (POROS), were evaluated on a 20-mer primer (5′-AACTTCATCCACGTTCACCT-3′, 10 μM). In addition to the unremoved Na+ and K+ in the primer solution, 5 mM MgCl2 was added to mimic the salt content in the primer extension reaction. Figure 2 shows the mass spectra of a salt-contaminated sample and of samples desalted by three different procedures. The effectiveness of the desalting methods was judged based on disappearance of salt-adducted peaks, improvement in ion intensity, and increase in signal-to-noise ratio. The results demonstrate varying degrees of desalting in the following order: POROS desalting > cationic exchange resin > ethanol precipitation. As shown in Fig.2a, the mass spectrum obtained from the unpurified primer displays a number of peaks representing a series of adducts formed with the Mg2+, Na+ and K+ ions. Compared to undesalted primer, the POROS desalting method (Fig.2d) shows the best desalting effect with respect to improved average intensity and signal-to-noise ratio (12 and 28 times, respectively). Furthermore, a combination of POROS and cationic exchange resin provides the optimal desalting effect. Fig.2e demonstrates the almost exclusively molecular ions of the 20-mer primer and the complete disappearance of salt adducts. This dramatic desalting effect resulted in a 120-times improved signal-to-noise ratio relative to the salt-contaminated starting material. Most importantly, high recovery was achieved during the sequential desalting, as seen by the elevated ion intensity in Fig.2e compared with that with either cation exchange resin or POROS R2 alone (Fig.2b–d). Based on these results, sequential desalting on POROS and cation exchange was incorporated into our multiplex genotyping strategy for common HBB alleles. Scheme 1 demonstrated the overall algorithm from DNA sample extraction to MALDI-TOF MS analysis.
Multiplex MS genotyping for HBB alleles
Parallel minisequencing reactions using PinPoint assays were evaluated for genotyping the eight common HBB alleles in the Taiwanese population. The typical mass spectrum of minisequencing primers plus the extended ddNTPs for wild types and carriers are shown in Fig.3. Primer extension products templated on the wild-type or mutant allele have the same length, and their mass difference is due to sequence difference at the polymorphic site (Table 1). Figure 3c and g show analysis of CD71/72 +A and CD17 A > T containing samples; although the mutant products partially overlap with one another because the mass difference is only 9 Da (the difference between A and T), the samples migrate at m/z=7929.1 and 6268.1, respectively, providing sufficient resolution for accurate genotyping.
MALDI-TOF MS analysis of eight human β-globin (HBB) gene alleles. For each SNP, the top, middle, and bottom panels show the mass spectra of primer, extension products of heterozygous carrier, and wild-type subject, respectively. a IVS2+654 C > T, b CD41/42 –TCTT, c CD71/72 +A, d CD1 T > G, e CD27/28 +C, f CD43 G > T, g CD17 A > T, and h −28 A > G. All panels: filled triangles and circles indicate primer-extension products from the wild-type and mutant subjects, respectively
For maximal benefit and clinical application, this method should differentiate homozygotes and heterozygotes carrying all eight common HBB mutations. Thus, we next performed the one-spectrum analysis on subjects in different genetic states, including a homozygous normal control, a carrier with a SNP (CD41/42 –TCTT), and mixture containing eight heterozygous carriers, as shown in Fig. 4. Because of the proximity of the two pairs of mutation sites (CD41/42 –TCTT and CD43 G > T, CD 27/28 +C and CD17 A > T), the eight minisequencing reactions were performed in parallel followed by the multiplex MALDI-TOF MS assay to overcome the obstruction caused by partially overlapping primers. The extension products from the eight parallel minisequencings were pooled for subsequent sequential desalting to reduce sample loss during purification as well as to speed up the procedure. As shown in Fig.4b and c, the eight pairs of primer/extended products for either wild-type or heterozygous carrier are well separated in one-base increments, which facilitate unambiguous data interpretation. Figure 4d shows assay results with the most complex input material, a mixture of eight heterozygous samples. In this assay, peaks for mutant and wild-type product were resolved for all eight SNPs, demonstrating that heterozygous carriers can be accurately genotyped by this method. This result demonstrated that parallel minisequencing followed by multiplex MALDI-TOF assay in combination with the mass tuning strategy is sufficiently accurate for the eight-fold multiplex genotyping diagnosis in β-globin gene.
Representative mass spectra from eight parallel PinPoint minisequencing followed by multiplex MALDI-TOF MS assay in the human β-globin (HBB) gene. a Control primers from minisequencing reactions. b Homozygous wild-type subjects. c Heterozygote with CD41/42 –TCTT mutation. d Mixure of eight heterozygous carriers. a–c: filled triangles and circles indicate primer-extension products from the wild-type and mutant subjects, respectively
In this study, a total of 80 subjects were analyzed, including 37 wild-type controls and 43 heterozygous cases (Table 2). All of the mutations can be identified without ambiguity, consistent with the characterization by direct sequencing. The highly accurate detection rate demonstrated the reproducibility of the approach for clinical diagnostics. Thus, the current approach is a reliable technique for mutation screening of β-thalassemia.
Diagnosis of HBB mutations in a core family
Ultimately, it would be useful to apply multiplex HBB genotyping for prenatal diagnosis. Here, we evaluated the potential of our assay for this application by analyzing HBB status in a nuclear family at risk for β-thalassemia. Figure 5a shows the MS analysis of all eight polymorphic sites. Seven normal allelles were detected, and the products of wild-type and mutant alleles corresponding to IVS2+654 C > T and CD41/42 –TCTT polymorphisms were detected. The mass spectra shown in Fig.5b are enlarged to facilitate analysis of the spectral region relevant to the genotype assignment. The spectra of parents (I and II) display characteristic heterozygous peaks consistent with the fact that they are heterozygous carriers for the mutation IVS2+654 C > T and CD41/42 –TCTT, respectively. For child III, the spectrum reveals a doublet peak at m/z=8648.7 and 8664.2 of mass difference 15 Da (Fig.5b), indicating that the IVS2+654 C > T allele was inherited from the father. Spectra for the other two children (IV and V) were identified as compound heterozygotes; they inherited both mutant alleles IVS2+654 C > T and CD41/42 –TCTT, and thus they are affected individuals and not carriers. These two mutations are the most commonly observed SNPs in the Taiwanese population (Su et al. 2003). In following with the Mendelian rules of inheritance, the multiplex genotyping assay confirmed that the offspring analyzed carried half of their alleles from their mother and half from their father. These data demonstrate that our multiplex genotyping strategy for common HBB alleles in the Taiwanese population is capable of genotyping heterozygote carriers and homozygote normal and affected individuals.
Genotyping of hereditary β-globin (HBB) gene mutations in a core family. a A representative mass spectra showing all eight polymorphic sites (father I). b Expanded mass spectra showing IVS2+654 C>T and CD41/42 –TCTT. Filled triangles and circles indicate primer-extension products from the wild-type and mutant subjects, respectively
Discussion
From a molecular genetics standpoint, β-thalassemia is a quite heterogeneous disease. Specific human populations at risk for β-thalassemia each carry a different subset of HBB alleles represented by a small number of common high-frequency mutations and several low-frequency mutations. Among the Chinese population in Mainland China, Taiwan, and North America, 27 alleles of HBB have been identified (Baysal and Carver 1995; Chiou et al. 1993; Kazazian 1990; Ko et al. 1997; Liang et al. 1994; Lin et al. 1991, 1992; Xu et al. 1996). However, in the more restricted Taiwanese population, only eight point mutations account for a great majority (98%) of β-thalassemia cases. Although multiplex minisequencing followed by multiplex analysis of sequencing products is a feasible genotyping approach for some genes (Meyer et al. 2004; Ross et al. 1998), technical difficulties have limited the general use of this method (Tost and Gut 2002). Because extension efficiency and optimum primer annealing temperature vary with the length and sequence of each primer (Syvanen 1999; Wang et al. 2003), it is difficult to optimize the multiplex minisequencing reaction (Butler et al. 2001).
The goal of this study was to devise a high-throughput genotyping assay for the eight common HBB alleles in the Taiwanese population. It was difficult to develop a multiplex sequencing strategy for these SNPs because of their proximity to one another, which leads to competition for primer-binding sites during the annealing step and reduces efficiency of the minisequencing reaction. The competition could be reduced by use of forward and reverse primers for the closely spaced alleles CD27/28 +C and CD17 A > T. However, this may reduce reaction efficiency by interfering with enzymatic extension or hybridization of some genotyping primers in the multiplex reaction (Ross et al. 1998). Low efficiency of primer extension could also be caused by off-target annealing (Su et al. 2003). This is evidenced in the case of CD41/42 –TCTT and CD43 G > T, which are only 5 bps apart, and their probing primers are partially overlapping (Fig.1). When used in multiplex primer extension reactions, the yield from these primers was low. We failed to generate efficient multiplex primer extension yields at these two target sites. To our knowledge, multiplex genotyping has never been used to detect the eight common HBB mutations among the general Taiwanese population. Considering these issues, an alternative approach was adopted in which eight parallel minisequencing reactions were performed, and the products of these reactions were pooled for subsequent analysis by MALDI-TOF MS. This strategy does not require time-consuming optimization and yet preserves the advantages of multiplexing by using pooled samples during desalting and MS analysis.
MALDI-TOF MS is an important technology characterized by highly accurate molecular mass measurement, high resolution, and coverage of a wide mass range. Because it is also suitable as a high-throughput assay, it is a promising alternative to conventional genotyping techniques. Considering the merits of multiplex genotyping using MALDI-TOF MS, the PinPoint assay provides several specific advantages. First, in the PinPoint assay, a primer is annealed to the targeted DNA immediately upstream of the SNP site and is extended by a single base in the presence of all four ddNTPs. Among all minisequencing methods, the PinPoint assay produces the smallest extension products, which are least susceptible to variation in extension efficiency (Haff and Smirnov 1997a, 1997b; Ross et al. 1998). Second, the primer and its extension product are in a narrow mass range. This is preferred in high-level MALDI-TOF-MS-based multiplex genotyping because many pairs of primer/extension products can be designed within a relatively narrow mass range, which facilitates the display and interpretation of multiple SNPs. Furthermore, all four dideoxynucleotides can be utilized in PinPoint assay without additional concerns of the different genotypes.
Metal cation with high affinity for the negatively charged sugar-phosphate backbone will cause reduced ion intensity and ambiguous mass assignments. Thus, multiplex genotyping, which demands high resolution and sensitivity, requires stringent sample desalting prior to MALDI-TOF MS. Various desalting techniques have been described and discussed (Guo 1999; Nordhoff et al. 1996; Ragas et al. 2000). A comparative study performed by Ragas et al (2000) has revealed that cation-exchange resin beads have an effective desalting effect for lower molecular weight oligonucleotides while higher molecular weight oligonucleotides require the more time-consuming minidialysis. To systematically develop an effective sample purification method with minimal loss during desalting procedures, we investigated various methods. In the current study, the combination of the POROS material and the cationic exchange resin demonstrated superior desalting without recovery loss, thereby establishing satisfactory genotyping with a high level of accuracy. Furthermore, the additional desalting by cation exchange resin was found to be very efficient, even over short incubation times, which increased the procedure time by only a few minutes. Therefore, sequential desalting is amenable to automation, and the modified purification procedure can be adapted for high-throughput SNP genotyping.
When higher mass analytes are analyzed by MALDI-TOF, there is usually some loss of sensitivity (Yang et al. 2003), and higher mass analytes with similar m/z values are difficult to separate at a given spectra resolution. In multiplex genotyping, the “mass tuning” strategy can be used to optimize primer sequence and length, keeping primer/extension product pairs as close as possible in size while better separating the primer and extension products from other SNP sites. In the mass spectra shown here, primers and all dideoxy-extension products are resolved, including A/T heterozygotes CD71/72 +A and CD17 A > T. However, partial overlapping was observed in few cases (specifically IVS2+654 C > T and the CD71/72 +A), which arose because of insufficient peak resolution in the linear mode (typically around 900 for the MALDI-TOF MS used in this study). More complete peak separation can be achieved by using a higher voltage ion source or by including sugar additives for MALDI matrices (Shahgholi et al. 2001); however, these methods were not needed to interpret the spectra shown here. Additionally, although no particular effort was made to optimize the yield of primer extension, our results unambiguously genotyped the A/T heterozygous alleles due to the lack of interference of salt adducts by our modified sample preparation procedure (described above).
It should be pointed out that it may be feasible to detect more than eight HBB alleles using the multiplex genotyping strategy described here if additional primers are selected judiciously. Taking into account several factors such as ideal primer length and primer extension efficiency, the ideal mass range spans from m/z=5000 to 9000 Da (Tost and Gut 2002). As shown in Table 1, our assay resolves samples between m/z=5440 (shortest primer) and 8376 (longest extension product). Multiplex assay in a single MS reaction can further reduce the cost per genotype to approximately US $0.15, which decreases the diagnostic cost remarkably (Bray et al. 2001). In addition to cost reduction, the efficiency of the strategy can also be increased by automated purification, analysis, and/or data interpretation. Thus, the method described in this study is a promising tool for the efficient and economical diagnosis of family heredity using high-level multiplex genotyping. Theoretically, though our diagnosis may be extended to detect more mutation products with larger mass, the spectra resolution beyond m/z=9000 may not be enough to differentiate the highly complex genotyping.
In summary, a cost-effective, efficient, accurate, high-throughput, multiplex genotyping strategy was developed for eight common HBB alleles in the Taiwanese population. This method uses parallel minisequencing, multiplex sequential desalting, and MALDI-TOF MS, and it accurately identifies all relevant genotypes, including a mixture of eight heterozygous carriers. Our result accurately displayed the hereditary map of parents and their offspring at the molecular level. The strategy described here can be adapted for genotyping other disease-causing mutations; as such, it may be widely applicable in the clinical setting for diagnostic genotyping.
References
Abdullah WA, Jamaluddin NB, Kham SK, Tan JA (1996) The spectrum of beta-thalassemia mutations in Malays in Singapore and Kelantan. Southeast Asian J Trop Med Public Health 27:164–168
Baysal E, Carver MF (1995) The beta- and delta-thalassemia repository, 8th edn. Hemoglobin 19:213–236
Bleicher K, Bayer E (1994) Various factors influencing the signal intensity of oligonucleotides in electrospray mass spectrometry. Biol Mass Spectrom 23:320–322
Braun A, Little DP, Koster H (1997) Detecting CFTR gene mutations by using primer oligo base extension and mass spectrometry. Clin Chem 43:1151–1158
Bray MS, Boerwinkle E, Doris PA (2001) High-throughput multiplex SNP genotyping with MALDI-TOF mass spectrometry: practice, problems and promise. Hum Mutat 17:296–304
Butler JM, Ruitberg CM, Vallone PM (2001) Capillary electrophoresis as a tool for optimization of multiplex PCR reactions. Fresenius J Anal Chem 369:200–205
Chiou SS, Chang TT, Chen PH, Lee LS, Chen TS, Chang JG (1993) Molecular basis and haematological characterization of beta-thalassaemia major in Taiwan, with a mutation of IVS-13′ end TAG–>GAG in a Chinese patient. Br J Haematol 83:112–117
Fei Z, Ono T, Smith LM (1998) MALDI-TOF mass spectrometric typing of single nucleotide polymorphisms with mass-tagged ddNTPs. Nucleic Acids Res 26:2827–2828
Foglieni B, Cremonesi L, Travi M, Ravani A, Giambona A, Rosatelli MC, Perra C, Fortina P, Ferrari M (2004) Beta-thalassemia microelectronic chip: a fast and accurate method for mutation detection. Clin Chem 50:73–79
Fortina P, Dotti G, Conant R, Monokian G, Parrella T, Hitchcock W, Rappaport E, Schwartz E, Surrey S (1992) Detection of the most common mutations causing beta-thalassemia in Mediterraneans using a multiplex amplification refractory mutation system (MARMS). PCR Meth Appl 2:163–166
Guo B (1999) Mass spectrometry in DNA analysis. Anal Chem 71:333R–337R
Haff LA, Smirnov IP (1997a) Multiplex genotyping of PCR products with MassTag-labeled primers. Nucleic Acids Res 25:3749–3750
Haff LA, Smirnov IP (1997b) Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res 7:378–388
Harksen A, Ueland PM, Refsum H, Meyer K (1999) Four common mutations of the cystathionine beta-synthase gene detected by multiplex PCR and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem 45:1157–1161
Henthorn PS, Smithies O, Mager DL (1990) Molecular analysis of deletions in the human beta-globin gene cluster: deletion junctions and locations of breakpoints. Genomics 6:226–237
Kazazian HH Jr (1990) The thalassemia syndromes: molecular basis and prenatal diagnosis in 1990. Semin Hematol 27:209–228
Kim S, Edwards JR, Deng L, Chung W, Ju J (2002) Solid phase capturable dideoxynucleotides for multiplex genotyping using mass spectrometry. Nucleic Acids Res 30:e85
Ko TM, Xu X (1998) Molecular study and prenatal diagnosis of alpha-thalassemias and beta-thalassemias in Chinese. J Formos Med Assoc 97:5–15
Ko TM, Tseng LH, Hsu PM, Guu IJ, Lin YW, Li SF, Lee TY, Chuang SM (1997) Molecular characterization of beta-thalassemia in Taiwan and the identification of two new mutations. Hemoglobin 21:131–142
Kurg A, Tonisson N, Georgiou I, Shumaker J, Tollett J, Metspalu A (2000) Arrayed primer extension: solid-phase four-color DNA resequencing and mutation detection technology. Genet Test 4:1–7
Li S, Haces A, Stupar L, Gebeyehu G, Pless RC (1993) Elimination of band compression in sequencing gels by the use of N4-methyl-2′-deoxycytidine 5′-triphosphate. Nucleic Acids Res 21:2709–2714
Li J, Butler JM, Tan Y, Lin H, Royer S, Ohler L, Shaler TA, Hunter JM, Pollart DJ, Monforte JA, Becker CH (1999) Single nucleotide polymorphism determination using primer extension and time-of-flight mass spectrometry. Electrophoresis 20:1258–1265
Liang R, Liang S, Jiang NH, Wen XJ, Zhao JB, Nechtman JF, Stoming TA, Huisman TH (1994) Alpha and beta thalassaemia among Chinese children in Guangxi Province, PR China: molecular and haematological characterization. Br J Haematol 86:351–354
Lin LI, Lin KS, Lin KH, Chang HC (1991) The spectrum of beta-thalassemia mutations in Taiwan: identification of a novel frameshift mutation. Am J Hum Genet 48:809–812
Lin LI, Lin KS, Lin KH, Cheng TY (1992) A novel −32 (C-A) mutant identified in amplified genomic DNA of a Chinese beta-thalassemic patient. Am J Hum Genet 50:237–238
Marvin LF, Roberts MA, Fay LB (2003) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin Chim Acta 337:11–21
Meyer K, Fredriksen A, Ueland PM (2004) High-level multiplex genotyping of polymorphisms involved in folate or homocysteine metabolism by matrix-assisted laser desorption/ionization mass spectrometry. Clin Chem 50:391–402
Nakai K, Habano W, Fujita T, Schnackenberg J, Kawazoe K, Suwabe A, Itoh C (2002) Highly multiplexed genotyping of coronary artery disease-associated SNPs using MALDI-TOF mass spectrometry. Hum Mutat 20:133–138
Nordhoff E, Kirpekar F, Roepstorff P (1996) Mass spectrometry of nucleic acids. Mass Spectrom Rev 15:67–138
Paracchini S, Arredi B, Chalk R, Tyler-Smith C (2002) Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res 30:e27
Ragas JA, Simmons TA, Limbach PA (2000) A comparative study on methods of optimal sample preparation for the analysis of oligonucleotides by matrix-assisted laser desorption/ionization mass spectrometry. Analyst 125:575–581
Ross P, Hall L, Smirnov I, Haff L (1998) High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol 16:1347–1351
Shahgholi M, Garcia BA, Chiu NH, Heaney PJ, Tang K (2001) Sugar additives for MALDI matrices improve signal allowing the smallest nucleotide change (A:T) in a DNA sequence to be resolved. Nucleic Acids Res 29:E91
Sherlock J, Cirigliano V, Petrou M, Tutschek B, Adinolfi M (1998) Assessment of diagnostic quantitative fluorescent multiplex polymerase chain reaction assays performed on single cells. Ann Hum Genet 62(Pt 1):9–23
Southern EM, Case-Green SC, Elder JK, Johnson M, Mir KU, Wang L, Williams JC (1994) Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res 22:1368–1373
Su YN, Lee CN, Hung CC, Chen CA, Cheng WF, Tsao PN, Yu CL, Hsieh FJ (2003) Rapid detection of beta-globin gene (HBB) mutations coupling heteroduplex and primer-extension analysis by DHPLC. Hum Mutat 22:326–336
Sun X, Ding H, Hung K, Guo B (2000) A new MALDI-TOF based mini-sequencing assay for genotyping of SNPS. Nucleic Acids Res 28:E68
Syvanen AC (1999) From gels to chips: “minisequencing” primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum Mutat 13:1–10
Tan KL, Tan JA, Wong YC, Wee YC, Thong MK, Yap SF (2001) Combine-ARMS: a rapid and cost-effective protocol for molecular characterization of beta-thalassemia in Malaysia. Genet Test 5:17–22
Thein SL, Winichagoon P, Hesketh C, Best S, Fucharoen S, Wasi P, Weatherall DJ (1990) The molecular basis of beta-thalassemia in Thailand: application to prenatal diagnosis. Am J Hum Genet 47:369–375
Tost J, Gut IG (2002) Genotyping single nucleotide polymorphisms by mass spectrometry. Mass Spectrom Rev 21:388–418
Ugozzoli LA, Lowery JD, Reyes AA, Lin CI, Re A, Locati F, Galanello R, Macioni L, Maggio A, Giambona A, Loutradi A, Boussiou M, Wallace RB (1998) Evaluation of the BeTha gene 1 kit for the qualitative detection of the eight most common Mediterranean beta-thalassemia mutations. Am J Hematol 59:214–222
Van Ausdall DA, Marshall WS (1998) Automated high-throughput mass spectrometric analysis of synthetic oligonucleotides. Anal Biochem 256:220–228
Vrettou C, Traeger-Synodinos J, Tzetis M, Palmer G, Sofocleous C, Kanavakis E (2004) Real-time PCR for single-cell genotyping in sickle cell and thalassemia syndromes as a rapid, accurate, reliable, and widely applicable protocol for preimplantation genetic diagnosis. Hum Mutat 23:513–521
Wang W, Kham SK, Yeo GH, Quah TC, Chong SS (2003) Multiplex minisequencing screen for common Southeast Asian and Indian beta-thalassemia mutations. Clin Chem 49:209–218
Weatherall DJ, Clegg JB (1996) Thalassemia—a global public health problem. Nat Med 2:847–849
Weatherall DJ, Clegg JB (2001) The thalassemia syndormes, 4th edn.
Wise CA, Paris M, Morar B, Wang W, Kalaydjieva L, Bittles AH (2003) A standard protocol for single nucleotide primer extension in the human genome using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 17:1195–1202
Wu G, Hua L, Zhu J, Mo QH, Xu XM (2003) Rapid, accurate genotyping of beta-thalassaemia mutations using a novel multiplex primer extension/denaturing high-performance liquid chromatography assay. Br J Haematol 122:311–316
Xu X, Liao C, Liu Z, Huang Y, Zhang J, Li J, Peng Z, Qiu L, Xu Q (1996) Antenatal screening and fetal diagnosis of beta-thalassemia in a Chinese population: prevalence of the beta-thalassemia trait in the Guangzhou area of China. Hum Genet 98:199–202
Yang H, Wang H, Wang J, Cai Y, Zhou G, He F, Qian X (2003) Multiplex single-nucleotide polymorphism genotyping by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem 314:54–62
Yip SP, Pun SF, Leung KH, Lee SY (2003) Rapid, simultaneous genotyping of five common Southeast Asian beta-thalassemia mutations by multiplex minisequencing and denaturing HPLC. Clin Chem 49:1656–1659
Acknowledgements
Financial support from Academia Sinica and the National Science Council, Taiwan of the Republic of China (NSC 92-2113-M-001-071) is gratefully acknowledged. Hsin-Kai Liao and Yi-Ning Su contributed equally to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liao, HK., Su, YN., Kao, HY. et al. Parallel minisequencing followed by multiplex matrix-assisted laser desorption/ionization mass spectrometry assay for β-thalassemia mutations. J Hum Genet 50, 139–150 (2005). https://doi.org/10.1007/s10038-005-0234-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-005-0234-z
Keywords
This article is cited by
-
Target-allele-specific probe single-base extension (TASP-SBE): a novel MALDI–TOF–MS strategy for multi-variants analysis and its application in simultaneous detection of α-/β-thalassemia mutations
Human Genetics (2023)
-
Comparison of the mismatch-specific endonuclease method and denaturing high-performance liquid chromatography for the identification of HBB gene mutations
BMC Biotechnology (2008)