Abstract
The 16p13.3 breakpoints of two de novo translocations of chromosome 16, t(1;16) and t(14;16), were shown by initial mapping studies to have physically adjacent breakpoints. The translocations were ascertained in patients with abnormal phenotypes characterized by predominant epilepsy in one patient and mental retardation in the other. Distamycin/DAPI banding showed that the chromosome 1 breakpoint of the t(1;16) was in the pericentric heterochromatin therefore restricting potential gene disruption to the 16p13.3 breakpoint. The breakpoints of the two translocations were localized to a region of 3.5 and 115 kb respectively and were approximately 900 kb apart. The mapping was confirmed by fluorescence in situ hybridization (FISH) of clones that spanned the breakpoints to metaphase spreads derived from the patients. The mapping data showed both translocations disrupted the ataxin-2-binding protein 1 (A2BP1) gene that encompasses a large genomic region of 1.7 Mb. A2BP1 encodes a protein that is known to interact with the spinocerebellar ataxia type 2 (SCA2) protein. It is proposed that disruption of the A2BP1 gene is a cause of the abnormal phenotype of the two patients. Ninety-six patients with sporadic epilepsy and 96 female patients with mental retardation were screened by SSCP for potential mutations of A2BP1. No mutations were found, suggesting that disruption of the A2BP1 gene is not a common cause of sporadic epilepsy or mental retardation.
Similar content being viewed by others
De novo chromosome rearrangements are associated with an increased risk of congenital malformations (Warburton 1991). Disruption of a critical gene at the translocation breakpoint is thought to be the basis for this increased risk. We report the finding of two patients with de novo translocations that involve a breakpoint at 16p13.3, show that there is disruption of the ataxin-2-binding protein 1 (A2BP1) gene by these 16p13.3 breakpoints, and propose that the disruption of A2BP1 is the basis for the patients’ clinical phenotypes.
The first of the de novo translocations was ascertained in a boy with severe intellectual and developmental retardation (developmental skills at 2–2.5 years when examined at 4 years 11 months) but with no dysmorphic features other than strabismus and down-turned angles of the mouth. There was a single recorded episode of fitting at 5 days old. Chromosome analysis showed a t(1;16)(q12;p13.3). Parental karyotypes were normal. Both parents were considered to have mild intellectual disability. Distamycin A/DAPI, which stains the pericentromeric heterochromatin of chromosomes 1 and 16, was used to stain the chromosomes (Fig. 1a). The der(1)t(1;16) showed a single band; the der(16) had two bands, one representing the 16 heterochromatin and the other at the 16p13.3 breakpoint derived from chromosome 1 heterochromatin (Fig. 1a). This observation is consistent with the location of the chromosome 1 breakpoint within the 1q12 heterochromatin. If the boy’s clinical anomalies result from disruption of a gene by the translocation, this gene must be on chromosome 16, since genes are absent in pericentromeric heterochromatin.
Mapping of translocation breakpoints. a Distamycin/DAPI banding of t(1;16) showing the chromosome 1 pericentromeric heterochromatin (indicated by arrows) and the chromosome 16 pericentromeric heterochromatin (indicated by asterisk). Both the der(1) and the der(16) have a chromosome 1 heterochromatin band and therefore the chromosome 1 breakpoint is in 1q12. bFluorescence in situ hybridization (FISH) of PAC 978I1 to metaphase chromosomes of t(1;16). Signal on the intact chromosome 16 and both the der(1) and der(16) show that this clone spans the chromosome 16 breakpoint. c Chromosome painting of metaphase spread of t(14;16) with chromosome 16 paint. d FISH of BAC 545E8 to metaphase chromosomes of t(14;16). Signal on the intact chromosome 16 and both the der(14) and der(16) show that this clone spans the chromosome 16 breakpoint
The second de novo translocation with a breakpoint in the same vicinity of 16p13.3 was a complex three-break rearrangement, t(14;16)(q32; p13.3) inv 16(p13.3 p12.1) (Callen et al. 2002). Chromosome 16 is visualized in this translocation in Fig. 1c by hybridizing a chromosome 16 paint to metaphase spreads from the patient. The boy with this translocation had mild dysmorphism, klinodactyly of the fifth finger, and mild cutaneous bilateral syndactyly of toes two and three. Assessment at 14 years was of mild mental retardation with a history of slow milestones and behavioral problems. A grand mal seizure at 5 years of age was followed by another 2 years later. There were two additional seizures and short absences before control with drug treatment was successful. Parental chromosomes were normal.
Physical mapping of the chromosome 16 breakpoints in these two patients was facilitated by the construction of mouse/human somatic-cell hybrids that retained the der(16) of the translocation in the absence of any other region of chromosome 16. The mouse/human somatic-cell hybrid CY196 retained the der(16)t(1;16), and the hybrid CY182 retained the der(16)t(14;16). These two hybrids enabled rapid positioning of the breakpoints at 16p13.3 within the established physical map of chromosome 16 that was based on an extensive somatic hybrid mouse/human hybrid panel (Callen et al. 1995). The available gene and DNA markers that were mapped to this panel could not separate the chromosome 16 breakpoints of the two hybrids, and therefore, more detailed mapping was undertaken.
DNAs from the somatic-cell hybrids CY182 and CY196 were used to construct a physical map of microsatellite and other PCR-based markers in the immediate vicinity of the breakpoints at 16p13.3 (Fig. 2). FISH to metaphase chromosomes of the translocations was used to detect genomic clones that bridged the translocation breakpoints. The FISH techniques and procedures used were as previously described (Callen et al. 1990). FISH of the YAC 761C2 showed that it spanned the 16p13.3 breakpoint of the t(1;16). This allowed the identification of the PAC clone 978I1 that also spanned the breakpoint, since a FISH signal was seen on both the der(1) and der(16) of the translocation (Fig. 1b). The DNA sequence of an overlapping BAC 578P21 was used to design oligo primers that were used to further localize the breakpoint by PCR analysis of DNA from the corresponding somatic-cell hybrid CY196. The breakpoint of CY196 was finally localized to a 3.46-kb region between bases 70,910 and 74,366 of BAC578P21.
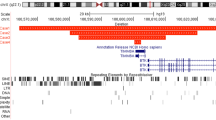
Physical map of 16p13.3 in the vicinity of the t(1;16) and t(14;16) showing the genomic structure of the A2BP1 gene. The contig of overlapping BAC clones is shown, together with the location of the YAC 761C1. Locations of the 16 exons of A2BP1 are shown as solid rectangles. Positions of the two de novo translocation breakpoints contained in the somatic-cell hybrids CY196 and CY182 are indicated by arrows
Physical mapping of the 16p13.3 breakpoint of the t(14;16) inv(16) required the construction of a contig of large insert clones. Start points for this physical map were previously mapped probes (cosmid c62F6, Callen et al. 1995) and additional microsatellite markers derived from public database (D16S3088, A008547 and WI-6421). A combination of in silico screening of the nr and htgs NCBI databases and screening of high-density BAC filters (RPCI-11 library, PAC/BAC Resources, Buffalo, NY) enabled identification of a contig that encompassed the 16p13.3 breakpoint. The contig was confirmed by restriction analysis using NotI and EagI and using probes generated from the ends of clones or from markers known to map in the region (data not shown). FISH to metaphase chromosomes of the t(14;16) translocation established that the breakpoint was contained within the BAC 545E8 (Fig. 1d). Analysis of the BAC 545E8 sequence showed that previously mapped PCR-based markers were present in this sequence, and this allowed the breakpoint to be restricted to a 115-kb region of the BAC. The chromosome 16 breakpoint of the t(1;16) is approximately 6 Mb from the telomere of the short arm of chromosome 16, and the 16p13.3 breakpoint of the t(14;16) is approximately 900 kb further proximal.
An in silico search was undertaken to identify genes in the vicinity of the two breakpoints at 16p13.3. Interestingly, gene A2BP1, a 2.3-kb transcript of 16 exons coding for a 377 amino acid protein, was found to encompass a very large genomic region of 1.7 Mb that included both chromosome 16 translocation breakpoints of the two patients (Fig. 2). The t(1;16) translocation removes the first large exon of A2BP1. Although the start codon of the open reading frame is in exon 4, the absence of the promoter and the majority of the 5′ untranslated region would make transcription from this truncated version of A2BP1 unlikely. It should also be noted that this translocation results in fusion of 16p13.3 with the centric heterochromatin of chromosome 1, and therefore it is unlikely that the translocation will generate any fortuitous juxtaposition of elements that could result in expression of A2BP1. The t(14;16) removes the first three exons of A2BP1 and also does not interrupt the open reading frame. Removal of the promoter and 5′ untranslated region would be expected to result in a lack of expression of this gene, since the breakpoint is in the third intron several hundred kb upstream of exon 4. It is possible, but unlikely, that the juxtaposed region from chromosome 14 may be compatible with transcription of A2BP1. It should be noted that the der(16) of the t(14;16) also possessed a small paracentric inversion of the short arm (Callen et al. 2002). There is the possibility that these additional breakpoints also disrupted genes that contributed to the patients’ clinical phenotype.
The A2BP1 gene was originally identified from a yeast two hybrid analysis, as the encoded protein interacts with the spinocerebellar ataxia type 2 (SCA2) protein (Shibata et al. 2000). A2BP1 has a predicted role as an RNA binding protein, and studies of the mouse ortholog confirm expression in the adult nervous system but also suggest a role in embryogenesis (Kiehl et al. 2001). The phenotypes of the two patients with disruption of A2BP1 are consistent with the gene’s expression and possible functional role. We attempted to compare the expression of A2BP1 by RT-PCR in the two patients with chromosome 16 translocations with the expression in normal individuals. Unfortunately, this was not possible, since there was no significant expression of A2BP1 in fibroblasts, the only patient material that was available for analysis. It is possible that mutations of A2BP1 may be a cause of sporadic autosomal dominant mental retardation or epilepsy. Accordingly, the exons corresponding to the open reading frame of A2BP1 were screened by single-strand polymorphism analysis in 96 female patients with mental retardation and 96 patients with idiopathic generalized epilepsy. The sequences of oligo primers for PCR amplification were homologous to the intron sequence flanking the exons (sequence available on request). Sequence variation was detected in the flanking intronic sequences of exons 2, 6, 12, the alternatively spliced 13A, and 15. The only variations detected within an exon were in exon 6, and these did not result in an amino acid change (see Table 1). These data suggest that mutations of A2BP1 are unlikely to contribute significantly to the etiology of sporadic mental retardation or epilepsy. It should be noted that heterozygous deletions of entire exons would not be detected by the procedures used.
A total of six de novo translocations were represented in the total of 60 balanced chromosome 16 translocations that had breakpoints mapped on the somatic cell hybrid panel (Callen et al. 1995). Of the six, in two cases, the subjects of this report were the only patients with known abnormalities. The breakpoints of these two translocations were within 300 kb, while the total length of chromosome 16 is approximately 100 Mb. It is unlikely that this is a chance finding. The association of phenotypic abnormality with disruption of an autosomal gene requires either a dominant-negative effect or haplo insufficiency, and this would be expected to be uncommon. We therefore speculate that the basis of the abnormal phenotypes in these two patients is disruption of the A2BP1 gene by the chromosome 16 breakpoints of their de novo translocations.
References
Callen DF, Baker E, Eyre HJ, Chernos JE, Bell JA, Sutherland GR (1990) Reassessment of two apparent deletions of chromosome 16p to an ins(11;16) and a t(1;16) by chromosome painting. Ann Genet 33:219–221
Callen DF, Lane SA, Kozman H, Kremmidiotis G, Whitmore SA, Lowenstein M, Doggett NA, Kenmochi N, Page DC, Maglott DR et al (1995) Integration of transcript and genetic maps of chromosome 16 at near-1-Mb resolution: demonstration of a “hot spot” for recombination at 16p12. Genomics 29:503–511
Callen DF, Eyre H, McDonnell S, Schuffenhauer S, Bhalla K (2002) A complex rearrangement involving simultaneous translocation and inversion is associated with a change in chromatin compaction. Chromosoma 111:170–175
Kiehl TR, Shibata H, Vo T, Huynh DP, Pulst SM (2001) Identification and expression of a mouse ortholog of A2BP1. Mamm Genome 12:595–601
Shibata H, Huynh DP, Pulst SM (2000) A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet 9:1303–1313
Warburton D (1991) De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet 49:995–1013
Acknowledgments
The National Health and Medical Research Council funded this research. We would like to thank Raman Kumar and Ross McKirdy for performing the RT-PCR assays.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhalla, K., Phillips, H.A., Crawford, J. et al. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet 49, 308–311 (2004). https://doi.org/10.1007/s10038-004-0145-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-004-0145-4
Keywords
This article is cited by
-
Enhanced long-term potentiation and impaired learning in mice lacking alternative exon 33 of CaV1.2 calcium channel
Translational Psychiatry (2022)
-
Rett syndrome linked to defects in forming the MeCP2/Rbfox/LASR complex in mouse models
Nature Communications (2021)
-
In vitro dexamethasone treatment does not induce alternative ATM transcripts in cells from Ataxia–Telangiectasia patients
Scientific Reports (2020)
-
Concentration-dependent splicing is enabled by Rbfox motifs of intermediate affinity
Nature Structural & Molecular Biology (2020)
-
The effect of Rbfox2 modulation on retinal transcriptome and visual function
Scientific Reports (2020)