Abstract
Parietal foramina [PFM], inherited usually in an autosomal dominant mode, is an extremely rare developmental defect characterized by a symmetrical, oval hole in the parietal bone. It can be present as either an isolated or a syndromic feature. PFM types 1 and 2 (PFM1 and PFM2) have been found to be caused by mutations in the MSX2 and ALX4 genes, located to chromosomes 5 and 11, respectively. After exclusion of both the above loci in a large Chinese pedigree with autosomal dominant PFM, a genome-wide search revealed a linkage of the PFM to markers at the 4q21-q23 region. The maximum LOD score from two-point linkage analysis is 3.87 for marker D4S2961. Analysis of co-segregated haplotype localized the region to a 20-cM interval that flanks D4S392 and D4S2945. Therefore, we concluded that the PFM in the family is a new PFM locus. Although three genes, BMPR1B, PP1 and IBSP, are located to 4q21-q25 and their functions are related to bone morphogenesis, no mutations were identified by sequencing analysis of their exons.
Similar content being viewed by others
Introduction
Parietal foramina (PFM) [MIM 168500] is an extremely rare congenital developmental anomaly of the parietal bone. The hereditary mode is usually autosomal dominant. PFM is characterized by symmetrical, oval defects in the parietal bone, normally obliterated during the fifth fetal month. They are situated on each side of the sagittal suture and separated from each other by a narrow bridge of bone. The size of the opening decreases with age and considerable intrafamilial variability is observed. PFM is benign and asymptomatic in most circumstances. However, seizures, scalp defects, and structural and vascular malformations of the brain can also occur simultaneously (Wilkie et al. 2000). Available information showed that it occurs in Asians, Africans, Europeans and Native Americans (Little et al. 1990). Hereditary cranium bifidum is the same entity as symmetric PFM (Little et al. 1990; Zabek 1987). Pang and Lin (1982) observed that the bilateral defects in one child evolved from a single midline opening via median ossification. PFM can be present as an isolated defect, but it can also be one of the characteristics of different syndromes (Golabi et al. 1984; Young and Swift 1985; Shaffer et al. 1993; Potocki and Shaffer 1996; Bartsch et al. 1996).
Bartsch et al. (1996) identified a deletion in the short arm of chromosome 11p13-p11 in a contiguous gene syndrome with multiple exostoses, enlarged parietal foramina, craniofacial dysostosis, and mental retardation. Wu et al. (2000) found that ALX4 located at 11p11 was a candidate gene for the PFM in the 11p11.2 deletion syndrome, and mutations in ALX4 were identified for non-syndromic PFM, PFM type 1 (PFM1) (Wuyts et al. 2000a,2000b; Mavrogiannis et al.2001), while haploinsufficiency due to heterozygous mutations in the MSX2 homeobox gene located at 5q34-q35 could also cause PFM, PFM type 2 (PFM2) (Wilkie et al. 2000; Wuyts et al. 2000a,2000b; Cohen 2000).
Here we report a new locus for non-syndromic PFM in a large Chinese pedigree.
Subjects and methods
Pedigree assessment
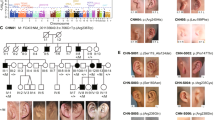
A large Chinese pedigree was identified through a proband who had typical features of PFM. All of the family members participated in the study and gave their informed consent. A total of 15 individuals (six males and nine females) in this four-generation family were affected with the defect. Nine of the affected and one normal individual were examined by cranial radiography. All the family members were carefully checked for multiple exostoses, craniofacial dysostosis, mental retardation, and other abnormal signs, but no syndromic forms were found. Hence, PFM was the only symptom found in the affected individuals of this family. All affected members had symmetrical, oval holes in their parietal bone.
Genotyping and linkage analysis
Samples of peripheral blood were taken from all available family members, and DNA was isolated by means of the laboratory procedure described elsewhere (Yang et al. 2000). First, a search was carried out on both chromosomes 11 and 5, because ALX4 was located on the former and MSX2 on the latter chromosome. However, no LOD score above 0 was obtained, so that a linkage was ruled out on these two chromosomes. A genome-wide screening was performed by a modified DNA-pooling method (Damji et al. 1998; Liu et al. 2001). A total of 383 autosomal microsatellite marker loci were genotyped in this family, using the ABI PRISM Linkage Mapping Set (version 2, LMS2) (Perkin-Elmer, USA). The set consists of fluorescence-labeled PCR primer pairs for 400 highly polymorphic dinucleotide-repeat microsatellite markers chosen from the Généthon human linkage map (Dib et al. 1996). These markers have an average spacing of 10 cM. Markers on the X chromosome were not used, because a male-to-male transmission was present in this pedigree.
Three pools of DNA from affected (n=14), unaffected family members (n=4) and unrelated population controls (n=6) were established by combining equal amounts of DNA from each individual. The control individuals were spouses of members of this pedigree. Multiplex PCR was performed for each panel of markers in a 5-μ l reaction volume containing 20 ng of pooling DNA, 3 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 200 μM each dNTP, 0.05 μM of each primer pairs and 0.5 U of Hot StarTaq polymerase (Perkin-Elmer). Reactions were performed on a Perkin-Elmer 9700 thermal cycler, and a touch-down thermocycling profile was used for all markers, consisting of an initial denaturation at 94 °C for 12 min, followed by 14 cycles each of denaturation at 94 °C for 30 s, annealing at 63 °C (decreased by 0.5 °C each cycle) for 90 s, and synthesis at 72 °C for 90 s. This was followed by 25 cycles, each of denaturation at 94 °C for 30 s, annealing at 56 °C for 90 s, synthesis at 72 °C for 90 s, followed by an extension step at 72 °C for 10 min, finally held at 4 °C. PCR products for each panel of markers were size fractionated on a 5% denaturing polyacrylamide gel by electrophoresis on an ABI 377 PRISMR DNA sequencer (Perkin-Elmer). The products were sized by the Genescan (Perkin-Elmer) version 2.1 program and scored by the Genotyper (Perkin-Elmer) version 2.0 program. Allele graphs showing a unique or higher peak in an affected panel lane after comparing with that of the unaffected and population control lanes (unaffected lane in particular) were regarded as candidate markers for typing of all members in this family. On chromosome 4, D4S1572 and D4S2964 showed a significantly higher peak; therefore, a total of 16 markers were selected for further mapping around the markers. Microsatellite markers, including D4S2945, D4S2961, D4S1591, D4S421 D4S2986, D4S1560, and D4S1559, were synthesized according to the Généthon human linkage map (Dib et al. 1996). PCR and thermocycling conditions were exactly the same as those used for the LMS2 marker set.
Two-point linkage analysis was performed by the MLINK program of the LINKAGE 5.1 package (Lathrop et al. 1985), based on a dominant inheritance model, with a disease penetrance of 0.95, and a disease gene frequency of 0.01. Allele frequencies and meiotic recombinations between the sexes were assumed to be equal. Multipoint linkage analysis was performed by the FASTMAP program (Curtis and Gurling 1993), with published inter-marker genetic distances from Généthon chromosome 4.
Results
Cranial X-ray photograph assessment
Nine of 15 individuals affected with PFM in this pedigree were available for cranial radiography. The result showed that these nine affected individuals had symmetrical, oval opening holes in their parietal bones (Fig. 1). Individual IV-1 carried a disease-associated haplotype, but her cranial X-ray showed no symmetrical holes on the parietal bone.
Linkage analysis
A total of 24 individuals available for genotyping were subjected to a chromosome-wide screening with all the markers from ABI LMS2 on chromosomes 5 and 11, since a linkage was previously identified on these chromosomal regions in non-sydromic PFM (Bartch et al., 1996; Wuyts et al.2000a,2000b. Unfortunately, no linkage (LOD score ≥0) was found. Therefore, both PFM1 and PFM2 loci were excluded from the disorder in our pedigree.
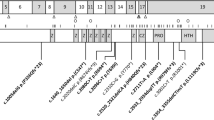
A genome-wide scanning on DNA pooling samples showed that a total of 95 markers were unique or higher in graphic peak in the DNA pooling sample of affected individuals after comparing electropherograms among three groups of pooling samples. These markers were selected for fine mapping. Eventually, only D4S1572 and D4S2964 were confirmed to be linked to the PFM phenotype, the remaining positive peaks being false positive. The maximum LOD score at θ=0.00 for the two markers were 3.29 and 1.70, respectively. Further linkage analysis was refined with 14 additional short tandem repeat (STR) markers spanning a 20-cM region around D4S1572. The result of the two-point LOD scores is summarized in Table 1. The highest two-point LOD score was 3.87 at D4S2961, whereas the multipoint LOD score reached the value of 6.17 between D4S2986 and D4S421 (Fig. 2). Subsequently, all of the remaining markers in the LMS2 set were typed for further verification, but no linkage (LOD score ≥0) on other loci was found. The results suggested that the disease in our family is a new PFM locus located to 4q21-q23.
Haplotype analysis
Extended haplotypes of the pedigree were originally constructed by the computer program Cyrillic (Cherwell Scientific, version 2.1.3) and then verified by the GENEHUNTER2.1 program by dividing the pedigree into three parts (Kruglyak and Lander 1998). The order of markers was based on the Généthon genetic linkage map: D4S405-D4S1592-D4S392-D4S2964-D4S1534-D4S414-D4S1559-D4S1560-D4S2986-D4S421-D4S1591-D4S1572-D4S2961-D4S2945-D4S406-D4S424-D4S413. The haplotypes were checked by view inspection, and a disease-linked haplotype was seen in all affected individuals (Fig. 3). However, an unaffected individual, IV-1, carried the whole disease-associated haplotype, and both III-8 and III-10 carried partial diseased-associated haplotypes as well. From these findings it is most likely that the disease locus is within a 20-cM interval, flanked by D4S2964 and D4S2961, on chromosome 4q21-q23 (Fig. 3).
Discussion
A total of 15 affected PFM individuals in this Chinese pedigree had typical clinical features, which are exactly the same as those reported previously (Fein and Brinker 1972; Okuda and Ikutomi 1965). The parietal hole is the only sign and all affected individuals had no other symptoms in this family except for III-3 and III-11, who developed recurrent headaches. This is probably due to their relatively large parietal holes, as a similar symptom reported by Pang and Lin (1982). Penetrance of PFM in this pedigree is incomplete. An unaffected individual, IV-1, carried the diseased haplotype, but she had no holes on her parietal bone. Both III-8 and III-10 carried partial disease-haplotypes. It remains to be investigated whether they are disease carriers with incomplete penetrance.
Muscle segment homeobox homologue 2 is an important target for the RAS signaling pathways and it establishes a balance between survival and apoptosis of neural crest-derived cells required for proper craniofacial morphogenesis. Mutations in this gene (MSX2) resulted in decreased parietal ossification (Cohen 2000). Heterozygous mutations or deletions in MSX2 have been found in patients with an isolated form of PFM from three unrelated British families (Wilkie et al. 2000). Similar mutations in MSX2 in four of their five PFM families were found by a Belgian research group (Wuyts et al. 2000a,2000b). They suggested that haploinsufficiency of MSX2 resulted in the skull defect. Later, in their remaining family, a mutation in the aristaless-like homeobox 4 gene (ALX4) was identified (Wuyts et al. 2000a,2000b). ALX4 encodes a paired-related homeodomain transcription factor, which decreases the expression of endogenous N-CAM protein (Boras et al 2002).
Another interesting finding was from a transgenic mouse model, where loss of MSX2 function could also induce holes in the mouse skull, but the location (frontal bone rather than the parietal bone) of the bony lesion is different from that of PFM in humans (Satokata et al. 2000). Discovery of the two different genes causing PFM suggested that different proteins might belong to the same or disparate pathological pathway; i.e., heterogeneity exists in PFM.
We failed to prove any linkages between PFM and markers on chromosomes 5 and 11 in this Chinese pedigree, but obtained a linkage to 4q21-q23. These findings indicate the presence of a third locus (PFM3) for PFM. Genes related to bone morphogenesis in this region may become potential candidate for the putative PFM3 gene. The bone morphogenetic protein receptor type IB gene (BMPR1B) encodes a member of the bone morphogenetic protein (BMP) receptor family of transmembrane serine/threonine kinases. BMPs are involved in endochondral bone formation and embryogenesis (Astrom et al 1999). The secreted phosphoprotein 1 gene (SPP1) encodes osteopontin, which is a bone and blood vessel extracellular matrix protein involved in calcification and atherosclerosis (Reinholt et al 1990). The integrin-binding sialoprotein gene (IBSP) encodes integrin-binding sialoprotein, which is a major structural protein of the bone matrix (Fisher et al 1990). Although we performed sequence analysis of these genes, we did not find any point mutations or deletions in their coding regions. It remains to be seen whether mutations exist in other candidate genes within the 4q21-q23 region.
References
Astrom AK, Jin D, Imamura T, Roijer E, Rosenzweig B, Miyazono K, ten Dijke P, Stenman G (1999) Chromosomal localization of three human genes encoding bone morphogenetic protein receptors. Mamm Genome 10:299–302
Bartsch O, Wuyts W, Van Hul W, Hecht JT, Meinecke P, Hogue D, Werner W, Zabel B, Hinkel GK, Powell CM, Shaffer LG, Willems PJ (1996) Delineation of a contiguous gene syndrome with multiple exostoses, enlarged parietal foramina, craniofacial dysostosis, and mental retardation, caused by deletions in the short arm of chromosome 11. Am J Hum Genet 58:734–742
Boras K, Hamel PA (2002) Alx4 binding to LEF-1 regulates N-CAM promoter activity. J Biol Chem 277:1120–1127
Cohen MM (2000) Craniofacial disorders caused by mutations in homeobox genes MSX1 and MSX2. J Craniofac Genet Dev Biol 20:19–25
Curtis D, Gurling H (1993) A procedure for combining two-point lod scores into a summary multipoint map. Hum Hered 43:173-185
Damji KF, Gallione CJ, Allingham RR, Slotterbeck B, Guttmacher AE, Pasyk KA, Vance JM, Pericak-Vance MA, Speer MC, Marchuk DA (1998) Quantitative DNA pooling to increase the efficiency of linkage analysis in autosomal dominant disease. Hum Genet 102:207–212
Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154
Fein JM, Brinker RA (1972) Evolution and significance of giant parietal foramina. Report of five cases in one family. J Neurosurg 37:487–492
Fisher LW, McBride OW, Termine JD, Young MF (1990) Human bone sialoprotein. Deduced protein sequence and chromosomal localization. J Biol Chem 265:2347-2351
Golabi M, Carey J, Hall BD (1984) Parietal foramina clavicular hypoplasia. An autosomal dominant syndrome. Am J Dis Child 138:596–599
Kruglyak L, Lander ES (1998) Faster Multipoint Linkage Analysis Using Fourier Transforms. J Comput Biol 5:1–7
Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37:482–498
Little BB, Knoll KA, Klein VR, Heller KB (1990) Hereditary cranium bifidum and symmetric parietal foramina are the same entity. Am J Med Genet 35:453–458
Liu W, Wang H, Zhao S, Zhao W, Bai S, Zhao Y, Xu S, Wu C, Huang W, Chen Z, Feng G, He L (2001) The novel gene locus for agenesis of permanent teeth (He-Zhao deficiency) maps to chromosome 10q11.2. J Dent Res 80:1716-1720
Mavrogiannis LA, Antonopoulou I, Baxova A, Kutilek S, Kim CA, Sugayama SM, Salamanca A, Wall SA, Morriss-Kay GM, Wilkie AO (2001) Haploinsufficiency of the human homeobox gene ALX4 causes skull ossification defects. Nat Genet 27:17–18
Okuda O, Ikutomi H (1965) Two cases of foramina parietalia permagna occurring in the same family. No To Shinkei 17:1255–1259
Pang D, Lin A (1982) Symptomatic large parietal foramina. Neurosurgery 11:33–37
Potocki L, Shaffer LG (1996) Interstitial deletion of 11 (p11.2p12): A newly described contiguous gene deletion syndrome involving the gene for hereditary multiple exostoses (EXT2). Am J Med Genet 62:319–325
Reinholt FP, Hultenby K, Oldberg A, Heinegard D (1990) Osteopontin — a possible anchor of osteoclasts to bone. Proc Nat Acad Sci USA 87:4473–4475
Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R (2000) MSX2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet 24: 391–395
Shaffer LG, Hecht JT, Ledbetter DH, Greenberg F (1993) Familial interstitial deletion 11(p11.12p12) associated with parietal foramina, brachymicrocephaly, and mental retardation. Am J Med Genet 45:581–583
Wilkie AO, Tang Z, Elanko N, Walsh S, Twigg SR, Hurst JA, Wall SA, Chrzanowska KH, Maxson RE (2000) Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat Genet 24:387–390
Wu YQ, Badano JL, McCaskill C, Vogel H, Potocki L, Shaffer LG (2000) Haploinsufficiency of ALX4 as a potential cause of parietal foramina in the 11p11.2 contiguous gene-deletion syndrome. Am J Hum Genet 67:1327–1332
Wuyts W, Cleiren E, Homfray T, Rasore QA, Vanhoenacker F, Van HW (2000a) The ALX4 homeobox gene is mutated in patients with ossification defects of the skull. J Med Genet 37:916–920
Wuyts W, Reardon W, Preis S, Homfray T, Rasore-Quartino A, Christians H, Willems PJ, Van HW (2000b) Identification of mutations in the MSX2 homeobox gene in families affected with foramina parietalia permagna. Hum Mol Genet 9:1251–1255
Yang X, She C, Guo J, Yu AC, Lu Y, Shi X, Feng G, He L (2000) A locus for brachydactyly type A-1 maps to chromosome 2q35-q36. Am J Hum Genet 66:892–903
Young ID, Swift PG (1985) Parietal foramina in the Saethre-Chotzen syndrome. J Med Genet 22:413–414
Zabek M (1987) Familial incidence of foramina parietalia permagna. Neurochirurgia 30:25–27
Acknowledgements
We would like to thank the National 973 and 863 Projects, the Shanghai Municipal Commission of Science and Technology, and the National Natural Science Foundation of China for generous financial support. We are also grateful to all of individuals of this family for their participation and to Dr. Wim Wuyts for providing valuable information concerning his research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, G., Zhang, D., Feng, G. et al. A novel locus for parietal foramina maps to chromosome 4q21-q23. J Hum Genet 48, 420–424 (2003). https://doi.org/10.1007/s10038-003-0050-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-003-0050-2
Keywords
This article is cited by
-
Enlarged parietal foramina: a rare forensic autopsy finding
International Journal of Legal Medicine (2016)
-
Enlarged parietal foramina: a review of genetics, prognosis, radiology, and treatment
Child's Nervous System (2013)
-
Enlarged parietal foramina caused by mutations in the homeobox genes ALX4 and MSX2: from genotype to phenotype
European Journal of Human Genetics (2006)