Abstract
The incidence of hepatoma is high in the Chinese population. Searching for genes involved in the functions of the liver, especially genes specifically expressed in the liver, will facilitate an insight into the molecular basis of normal and abnormal liver functions. Based on a differentially displayed cDNA fragment, which was down regulated in hepatoma tissues, we cloned a novel cDNA of 957 bp, TCP10L (T-complex protein 10 like), from the human liver cDNA library. Northern hybridization of this novel gene in 30 adult human tissues was examined. The result revealed that TCP10L expressed specifically in the human liver and testis. The TCP10L contains a 645-bp open reading frame encoding a deduced protein of 215 amino acids. As the deduced protein was analyzed further, a typical leucine zipper motif was found. We firstly examined the transcriptional function of the TCP10L protein by transfecting recombinant pM-TCP10L into mammalian cells. The subsequent analysis based on the dual luciferase assay system showed that TCP10L significantly inhibited the expression of reporter genes. Compared with that of the negative control, the luciferase activity were down regulated in HEK293 and SK-HEP-1, CHO cells by about 2.6, 9.8, and 5.5 folds respectively. A mutated type of TCP10L was also constructed. It showed that the repression of TCP10L to the expression of the reporter gene almost completely decreased, suggesting that the leucine zipper structure is critical for TCP10L to play its role in regulation function. Then we transfected the recombinant TCP10L-EGFP into cells. The results indicated that TCP10L subcellularly located in nuclei, either in HEK 293 or SK-HEP-1 cells. In addition, human TCP10L was found comprised of five exons and four introns, and mapped to chromosome 21q22.11.
Similar content being viewed by others
Introduction
The liver is involved in many important functions such as maintaining the metabolic balance of nutrients including fat, and carbohydrate; synthesizing substances, such as bile acid; and playing a central role in the detoxification of poisonous substances. In addition, the liver is also an important organ in the body’s defense mechanisms and a critical hematopoietic organ in the fetal stage (Petti et al. 1985; Houssaint et al. 1988). Liver diseases, especially hepatitis, cirrhosis, and hepatoma, are common in the Chinese population (Yeh et al. 1989; Lee 1983; Sun Z et al. 1991). Therefore, cloning and identification of the genes related to liver functions, especially those specifically expressed in the liver, might help us gain insights into the molecular mechanism of the development, physiological functions, and functional disorders of the liver.
In recent years, the gene expression profile in the human fetal liver has been investigated, and many tissue-specific and developmental-stage-specific genes in the liver were identified (Yu et al. 2001). For instance, cholesterol-7-hydroxylase NADPH: oxygen oxidoreductase (7-hydroxylase) is a liver-specific gene product that controls bile acid synthesis, the major pathway responsible for eliminating cholesterol from the body (Edwards et al. 1996). Besides, some researchers compared the gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver tissues, and a number of candidate genes altered in their expression levels have been found (Xu et al. 2001). Among these tissue-specifically-expressed genes, the genes encoding transcription factors may be of importance. Through regulating the expression of other genes, transcription factors can influence their target genes and subsequently influence the functions of tissues or organs. Therefore, studies on those transcription-factor genes specifically expressed in the liver may give us clues to the molecular basis of the liver’s physiologic functions.
In the present study, a novel gene, human TCP10-like gene, which is specifically expressed in the liver and testis, was identified. It was found that the TCP10-like gene is located in nuclei of mammalian cells and bears transcription inhibition activity. We propose that the TCP10-like gene might be a transcription factor gene and might have roles in the development of the liver.
Materials and methods
cDNA cloning and sequencing
In a differential display analysis between hepatoma tissues and their adjacent nontumor tissues from a female Chinese patient suffering from hepatocellular carcinoma, a 350-bp cDNA fragment down regulated in hepatoma tissues was found. Afterward, the cDNA fragment was sequenced and used as an information probe to search the human EST division in GenBank with the BLASTn program. A series of ESTs (AA758217, AA832234, AA927467, AI183985, AI004575, H94895, AA621014, AA621071, F11677, and AA563930) were obtained and assembled into a 957-bp EST contig. These published cDNA sequences provided the basis for designing the following two primers: JY-A, 5’-ATG ATC TGA TCC TGG CAG TAG TG −3’; and JY-B, 5’-CAG TGT AGA GTG ACA CAG GTG TC-3’). The primers were custom-made by ShengGong Inc (Shanghai, China) and were used to amplify the full-length cDNA from human λgt11 liver cDNA library (Clontech). PCR conditions were as follows: 1 µl template (>108 plaque-forming U/ml) was amplified in a final volume of 50 µl containing 5 µl of 10×PCR buffer, 1 µl of 20 mM dNTPs, 1.5 µl of 2.5 mM MgCl2, 2 U Taq polymerase (Promega) and 1 µl of 25 mM of each specific primer. PCR reactions were run on a PTC-200 DNA engine (MJ Research, Watertown, MA, USA) for 34 cycles (1 min at 94°C, 1 min at 61°C, and 1 min at 72°C) after an initial denaturation at 94°C for 3 min, and were followed by incubation at 72°C for 10 min. The amplification product was cloned into vector pGEM-T and five independent clones were sequenced using an ABI PRISM 377 DNA sequencer subsequently.
Plasmid construction and analysis of transcriptional function
The full-length ORF of the TCP10L cDNA was inserted in frame into mammalian cell expression plasmid pM (Clontech) with EcoRI/XbaI restriction enzyme sites to generate fusion genes encoding PM-TCP10L. The primers used in this construct were PM-TCP10LA 5’-GCG GAA TTC ATG CTG GCA GGT CAA CTC −3’, PM-TCP10LB 5’-GCG TCT AGA CCT TTC CAT CTT CAG AC-3’. The constructs were verified by DNA sequencing. Human HEK293, CHO, SK-Hep-1 cells were, respectively, grown in 24-well plates. The FuGENE transfection reagent (Roche) was used to perform the transfections as described in the manufacturer’s protocol. The recombinant PM-TCP10L, the empty pM, and the positive control pM3-VP16, pM-ZNF191 were cotransfected into cells respectively with reporter plasmid pGAL45tkLUC containing five consensus GAL4 binding sites and thymidine kinase minimal promoter upstream of the luciferase. We then used 0.6 µg of mixed plasmids in each experiment. The plasmid pRL-SV40 encoding Renilla luciferase was used as the internal control in each transfection. The day before transfection, about 2~6×104 cells were seeded per well of a 24-well plate in appropriate growth medium with serum. The cells were incubated at 37°C with 5% CO2.
Forty-eight hours after the transfection, the adherent cells were washed with 1× phosphate-buffered saline and lysed in 1 ml passive lysis buffer (Promega) The dual-luciferase reporter assay system ( Promega) was used to assay luciferase activity of the samples. The cell lysate was firstly assayed for the firefly luciferase activity using 100 µl of the substrate LARII. Then 100 µl of the Stop & Glo reagent was added to quench the firefly luciferase activity and activate the Renilla luciferase activity. Each set of transfections was repeated three times. The relative light units were measured by luminometer (Lumat LB9507).
Intracellular localization assay
Two primers were designed to amplify the TCP10L cDNA. EGFP-TCP10LA: 5’-CCC GAA TTC TCA TGC TGG CAG GTC-3’, EGFP-TCP10LB: 5’-ATA GGA TCC CGG ACA CCC CCC CG −3’. The acquired fragment digested with EcoRI/BamHI was inserted into the corresponding sites of enhanced green fluorescent protein (pEGFP-N1, Clontech) vector. Two lines of cells (HEK293 and SK-Hep-1) were seeded in 35-mm tissue culture plates and grown in suitable medium supplemented with 10% fetal bovine serum in a 5% CO2 incubator at 37°C. Cells were transfected the next day with EGFP-TCP10L using FuGENE transfection reagent (Roche) Each transfection was performed in duplicate. The transfected cells were grown for an additional 48 h and then examined using a Leica confocal microscope.
Northern blot analysis
Northern hybridizations were performed on multiple-tissue Northern membranes with mRNA samples from adult human tissues. The probes were prepared by labeling the cDNA fragments with [α-32P]dATP (Amersham) using the PCR method and purified on a Sepharose-G50 column. The membranes were prehybridized in hybridization/prehybridization solution {5×SSPE [1×SSPE = 0.15 M NaCl/10 mM sodium phosphate (pH 7.4)/1 mM EDTA], 50% formamide, 10×Denhart’s (1×Denhardt’s=0.02% Ficoll 400/0.02% polyvinylpyrrolidone/0.002% BSA), 2% (w/v) SDS, 100 µg/ml calf-thymus DNA} at 42 °C for 16 h, and hybridized in a hybridization oven (Hybaid Interactive, Teddington, Middx, UK) with the labeled probe for a further 24h continuous shaking. The membranes were then washed several times with 0.1×SSC (1×SSC=0.15 M NaCl/0.015 M sodium citrate) containing 0.1% SDS at 42°C and were exposed to X-ray film at −80°C for 5 days.
Results
Cloning of human TCP10L
Using the primers JY-A,B derived from the correlated EST sequences, we applied PCR to produce a 729-bp of JY-AB fragment from human λgt11 liver cDNA library. The automated sequencing showed that the sequences of all the clones in pGEM-T vector selected independently, are consistent with that of the EST contig (nt49–777), which we assembled formerly. Searching the GenBank nonredundancy database with the assembled 957-bp contig, it was found that this cDNA sequence had not been reported in the known databases. The gene was submitted to GenBank under the accession number of AF115967. We searched GenBank with the nucleotide sequence of the novel gene and found that this gene is similar to some members of human T-complex protein 10 (TCP10) gene family, such as TCP10A (U03399) (Islam et al. 1993). When human TCP10A was aligned with the novel putative protein, an identity about 39.7% was discovered in their amino acid sequences. We thereby submitted our gene to the HUGO gene nomenclature committee, and this novel gene was symbolized as TCP10L, TCP10-like gene.
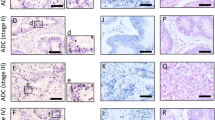
Using the ORF Finder analysis tool in NCBI, we found the TCP10L cDNA contains an intact open reading frame encoding a putative polypeptide of 215 amino acids (nt 88–735)(Fig. 1A). In the upstream of the initial codon ATG of TCP10L cDNA, there are three stop codons (TGA, nt 54–56; TAG, nt 67–69; TGA, nt70–72); while in its 3’-UTR, there are a typical polyadenylation signal (AATAAA, nt822–827) and an atypical one (ATTAAA, nt866–871). The deduced TCP10L is predicted to have an approximate molecular mass of 52KD and an acidic PI of 5.13 by ExPASy program (http://www.expasy.ch). Additionally, a leucine zipper motif was found near the middle part of TCP10L by 2ZIP program (http://www.dkfz-heidelberg.de/tbi/services/2zip) (Bornberg et al. 1998). The leucine zipper motif spans 25 amino acids from the residue 72–96, and contains four periodic repeats of leucine residues (residue 75, 82, 89, 96) (Fig. 1C). Moreover, the leucine zipper motif exists in a coiled coil conformation predicted by 2ZIP program.
The cDNA sequence, the putative protein, and the leucine zipper motif of human TCP10L. A The sequences of human TCP10L cDNA and its putative TCP10L protein. The sequences shaded in gray are the two primers, JY-A and JY-B, to amplify the full-length cDNA from the human λgt11 liver cDNA library; the letters underlined are the start codon, the asterisk indicates the stop codon; two poly (A) signal are boxed. In addition, the regular numbers are the numbers of the nucleotides, and the italic numbers are the numbers of amino acids. The GenBank accession numbers of the TCP10L gene is AF115967. B The structure models of the TCP10L protein and other 13 proteins containing the leucine zipper motif. The boxes are the position of the leucine zipper motifs in these proteins. The other 13 proteins are BACH2 (Genbank accession number: XM_031521), c-MYC (X00364), C/EBP (X52560), KRM12 (AF109780), LOC90183 (XM_029709), NRL (XM_033338), LAZipII (AF408398.1), c-FOS (V01512), GCN4 (K02205), TBZF (AB032478), BATF (XM_041063), MafK (AF059194), AP-1 (J04111). C Comparison of the leucine zipper motifs of the 14 proteins. All of the protein sequences are displayed in single-letter code. The abbreviations of Hs, Dr, Mm, Sc and Nt represent Homo sapiens, Danio rerio, Mus musculus, Saccharomyces cerevisiae, Nicotiana tobacum respectively. The digits shaded in gray represent the non-leucine-zipper region of the 13 proteins. The letters enlarged and bolded show the periodical arrays of leucine or methionine residues
To further explore the function of TCP10L, this leucine zipper motif was compared to the leucine zipper motifs in other 13 proteins containing leucine zipper (Fig. 1B and C), which are BATF (GenBank accession number: XM_041063), TBZF (AB032478), LAZipII (AF408398.1), MafK (AF059194), c-FOS (V01512), GCN4 (K02205), KRM12 (AF109780), AP-1 (J04111), NRL (XM_033338), LOC90183 (XM_029709), C/EBP (X52560), BACH2 (XM_031521), c-MYC (X00364). Structure comparison of these proteins revealed that the structure of the leucine zipper motif in TCP10L is reliable, although the position of the leucine zipper might appear near the N-terminus or the C-terminus or in the middle part of proteins (Fig. 1B). Besides, we still noticed that more than half the proteins (eight out of 14) have leucine zippers near C-termini. It is also observed that in leucine zippers, the number of leucine residues is variable from four repeats to six, which may govern stability and orientation of dimerization (Fig. 1C).
Assay of transcriptional function
Several protein motifs, which are essential to DNA recognition and binding, have been identified in transcription factors, such as the leucine zipper, zinc finger, helix-loop-helix, and helix-turn-helix (Busch et al. 1990). Accumulating data show that the leucine zipper motif, first postulated by Landschulz et al. (1988) , exists in numerous transcription factors such as c-Myc (protooncoproteins) (Amati et al. 1993; Luscher et al. 1999), AIF3, Jun and Fos (bZip family members of transcription factors) (Perez et al. 2001; Smith et al. 1990), and GCN4 (Sellers et al. 1989; Kouzarides et al. 1989). The leucine zipper is indispensable to the biological functions of these transcription factors through dimerization.
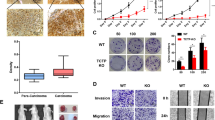
Accordingly, we speculated that the TCP10L might possess transcription-regulating activity because of its leucine zipper motif. To elucidate the significance of the leucine zipper in TCP10L, the experiment based on the dual luciferase assay system was performed to assay the transcription function. The ORF of TCP10L was inserted into mammalian cell expression plasmid pM, and the recombinant plasmid pM-TCP10L was constructed to express the fusion protein Gal4-TCP10L. Plasmid pM-TCP10L was cotransfected into different cell lines with reporter plasmid pGAL45tkLUC encoding firefly luciferase and the plasmid pRL-SV40 encoding Renilla luciferase, respectively.
One negative control, mock pM, and two positive control, pM3-VP16 and pM-ZNF191, were introduced in this experiment. Among them, VP16 is a known transactivator and ZNF191 is known as a transcriptional repressor (Han et al.1999). The luciferase activities of samples were determined by the Dual-Luciferase Assay System (Promega). Fig. 2 shows that recombinant TCP10L could significantly inhibit the expression of the reporter gene. Compared with that of the negative control, the luciferase activity were down-regulated by about 2.6, 9.8, and 5.5 folds in HEK293, SK-HEP-1, and CHO cells, respectively (p<0.001, n=4). As shown in Figs. 2B and C, the inhibition activity of TCP10L is much more remarkable in SK-HEP-1, a hepatoma-derived cell line, than in HEK293 cells.
Transcription function analysis of human TCP10L protein. By cotransfecting recombinant plasmids pM-TCP10L with reporter plasmid pGAL45tkLUC, determination of luciferase activity was performed in CHO (A), HEK293 (B), and SK-HEP-1 (C) cells respectively. One negative control, pM, and two positive controls (a transcriptional activator pM3-VP16, a transcriptional repressor pM-ZNF191) were introduced into this assay system. The plasmid pRL-SV40 encoding Renilla luciferase was used as the internal control in each transfection. D Dose-dependent inhibition of pM-TCP10L to the activity of the reporter gene pGAL45tkLUC. A fixed amount of reporter gene with varying amounts of TCP10L was used; the total amount of DNA was equalized by adding 10 ng pRL-SV40. E Dose-dependent inhibition of pM-ZNF191 to the activity of the reporter gene as control. A fixed amount of reporter gene with varying amount of ZNF191 was used. F Comparison of the luciferase activity of wild type TCP10L and its mutated type TCP10Lm in SK-HEP-1 cell line. The positive control, pM-ZNF191 is also shown. G The leucine zipper sequence of wild type TCP10L and its mutated type TCP10Lm, where the second leucine residue (UUG) of the zipper was replaced instead of phenylalanine (UUC). In this figure, the blank columns, the gray columns, the black columns, and the diagonal columns indicate transfection of pM, pM-TCP10L, pM3-VP16, and pM-ZNF191 respectively. Each value in this figure represents the mean of the enzyme activities determined in four assays. The error bars indicate standard deviation from means
Further, we examined whether transcription inhibition of the pGAL45tkLUC reporter gene activity was proportioned to the used amount of TCP10L. In this experiment, we used a fixed amount of pGAL45tkLUC reporter gene with varying amounts of TCP10L to transfect SK-HEP-1 cells, and the total amount of DNA was equalized by adding 10 ng pRL-SV40. In Figs. 2D and E, we showed that TCP10L gave dose-dependent expression inhibition of the pGAL45tkLUC reporter gene (p<0.001, n=4) and that ZNF191 as a control also gave dose-dependent repression to the expression of the reporter gene (p<0.001, n=4).
As we know, the leucine zipper consists of a periodic repetition of leucine residues at every seventh position, and regions containing them appear to span eight turns of alpha-helix. The leucine side chains that extend from one helix interact with those from a similar helix, hence facilitating dimerization in the form of a coiled-coil. Thus, the zipper is important for the protein to bind to a specific sequence of DNA. Therefore, we constructed a mutated type of TCP10L where the second leucine residue (UUG) of the zipper was replaced instead of phenylalanine (UUC) (Fig. 2G). It showed that the repression of TCP10L to the expression of the reporter gene almost completely decreased (p<0.001, n=4) (Fig. 2F). Another mutation, the third leucine residue (CUC) of the zipper replaced by arginine (CGC), also lead to the inhibitory activity loss of TCP10L (data not shown). The results suggest that the leucine zipper structure is critical for TCP10L to play its role in the regulatory function.
Subcellular localization of human TCP10L fused with EGFP
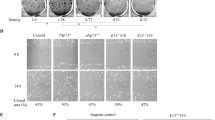
After we indicated that the TCP10L have a transcription repression function, we further explored the cellular localization of TCP10L. We hypothesize that, if TCP10L is a transcription factor, it must localize in nuclei. In order to test our assumption, we fused TCP10L with EGFP (enhanced green fluorescent protein) to explore its intracellular localization. When TCP10L-EGFP plasmid was transfected into cells, it showed that the fusion protein accumulated in nuclei both in the HEK293 and in SK-HEP-1 cell lines, while EGFP protein, the control, diffused all over the cell (Fig. 3). Thus, the nuclear accumulation experiments of TCP10L proved our hypothesis.
Tissue expression patterns of human TCP10L
In order to further explore TCP10L function, the expression pattern of human TCP10L in adult human tissue was also examined by Northern hybridization. Using isotope-labeled JY-AB fragment as a probe, the hybridization to the membrane with blotting mRNA from human tissues showed that a transcript about 1.0 kb abundantly expressed in the liver and testis, but cannot be found in 28 other tissues (Fig. 4). In the female, TCP10L only expresses in the liver. Metabolic reactions including synthesis and detoxification in human liver cells are very active, and many transcription factors are involved in the expression regulation of various genes participating in these reactions (Schiaffonati et al. 1997). We suggested TCP10L might be one of those candidates.
The expression pattern of the TCP10L in human tissues. A Multiple-tissue Northern blots (MTN I and MTN II, Clontech) containing total RNA from 16 adult human tissues were hybridized with the isotope-labeled TCP10L cDNA, and the β-actin was used as control. B MHB1004 and MHB1006 containing total RNA from 16 adult human tissues were hybridized with the isotope-labeled TCP10L cDNA, and the β-actin was used as control. With the Megaprime DNA labeling system (Amersham), the probes were prepared by labeling the cDNA fragments with [α-32P]dATP and were purified on a Sepharose-G50 column
Chromosome localization and genomic structure of human TCP10L
To understand the genomic characters of human TCP10L, we searched the on-line database in the Web site of the Human Genome Project Working Draft (http://genome.ucsc.edu), using the full length TCP10L cDNA as the query sequence. With the BLAT program (Kent 2002), the sequence of TCP10L was aligned to the sequences in the database, and the information of the localization and genomic structure of TCP10L was obtained. The result showed that TCP10L is localized at chromosome 21p22.11, and its genomic size is 8955 bp (Fig. 5A). As shown in Fig. 5B, human TCP10L comprises of five exons and four introns. The sizes of each intron and exon are shown in Fig. 5C. An STS marker, RH103054, is localized in the fifth exon, and another STS marker, SHGC-170092 is around exon 4 (Fig. 5B). In addition, we noticed that three genes, GCFC (AF153208), SYNJ1 (Cremona et al. 2000, Saito et al. 2001), and KIAA0539 (Nagase et al. 1998) are in the vicinity of the TCP10L and that two disease-related genes, SOD1 (Munch et al. 2001) and TIAM1 (Ives et al. 1998), have also been mapped to the same chromosome segment, 21p22.11.
Chromosome locus and genomic structure of the TCP10L. A Genomic localization of the TCP10L gene. The TCP10L gene is localized at chromosome21q22.11. Three genes, GCFC, SYNJ1, KIAA0539, which are adjacent to the TCP10L gene, are also shown. The hollow arrows represent the transcription direction of each gene. B Genomic organization of human TCP10L gene. It consists of five exons and four introns. The black blocks numbered and drawn to scale represent the exons. There are two STS markers, SHGC-170092 and RH103054, near the fourth and fifth exon. C The size and position of the exons of the TCP10L gene. The sequences flanking the exon/intron boundaries are given
Discussion
The liver is an essential organ bearing many physiological functions. Northern hybridization results in our study showed that TCP10L is a novel gene specifically expressed in the liver and testis. The gene structure showed that it was composed of five exons and four introns and spanned about 9.0 kb. Although the flanking sequence of the initial codon ATG is not completely consistent with Kozak rule, there are three stop codons before ATG, indicating the cDNA we cloned is nearly full length. The TCP10L contains the representative character of the leucine zipper motif, with a leucine residue in every seventh position. The leucine zipper motif has been observed in a number of proteins thought to function as eukaryotic transcription factors. We hypothesized human TCP10L is also a transcription factor. In order to know whether it is a transcriptional activating or repressing factor, we used the dual luciferase assay system. It showed that the TCP10L fusion gene inhibited significantly the expression of reporter plasmid pGAL45tkLUC in CHO, HEK293, SK-Hep-1 cell lines.
It is reported that the leucine residues could be replaced with Met or Ile or Val in some leucine zippers (Bornberg et al. 1998). Take human MafK(AF059194) protein as an example. There is a methionine residue among the periodical repeated leucine residues. In our experiment, when the critical leucine zipper motif in TCP10L was demolished by mutation, its transcriptional inhibition was almost disappearing. Our experimental results indicated that human TCP10L is a kind of transcription repressing factor. The subcellular localization that the gene is accumulated in the nuclei further proved our hypothesis to some extent.
In recent years, more and more researches have indicated that there are strong associations between oncogenesis and transcription factors possessing the leucine zipper motif (Amati et al. 1993, Luscher et al. 1999, Smith et al. 1990, Cremona et al. 2000). Based on the abundant expression of TCP10L in the liver and testis, we suggested that human TCP10L, with its transcription inhibition activity, might also play roles in the processes of cell proliferation and differentiation in the liver and testis. Further experiments are being done in our lab to prove our suggestion.
In conclusion, human TCP10L, a novel gene specifically expressed in the liver and testis, was cloned and characterized, and its protein product has been testified to have transcription inhibition activity. The present study is of potential value to help us understand the molecular mechanisms of liver physiological and pathological functions.
References
Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H (1993) Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72:233–245
Bornberg-Bauer E, Rivals E, Vingron M (1998) Computational approaches to identify leucine zippers. Nucleic Acids Res. 26:2740–2746
Busch SJ, Sassone-Corsi P (1990) Dimers, leucine zippers and DNA-binding domains. Trends Genet. 6:36–40
Cremona O, Nimmakayalu M, Haffner C, Bray-Ward P, Ward DC, De Camilli P (2000) Assignment of SYNJ1 to human chromosome 21q22.2 and Synj12 to the murine homologous region on chromosome 16C3–4 by in situ hybridization. Cytogenet Cell Genet. 88:89–90
Edwards PA, Davis RA (1996) Biochemistry of lipids, lipoproteins and membranes, 3rd edn. New Comprehensive Biochemistry 31, Elsevier, Amsterdam, pp 341–362
Han ZG, Zhang QH, Ye M, Kan LX, Gu BW, He KL, Shi SL, Zhou J, Fu G, Mao M, Chen SJ, Yu L, Chen Z (1999) Molecular cloning of six novel krüppel-like zinc finger genes from hematopoietic cells and identification of a novel transregulatory domain KRNB. J Biol Chem 274:35741–35748
Houssaint E, Hallet MM (1988) Inability of adult circulating haemopoietic stem cells to sustain haemopoiesis in mouse fetal liver microenvironment. Immunology 64:463–467
Islam SD, Pilder SH, Decker CL, Cebra-Thomas JA, Silver LM (1993).The human homolog of a candidate mouse t complex responder gene: conserved motifs and evolution with punctuated equilibria. Hum Mol Genet 2:2075–2079
Ives JH, Dagna-Bricarelli F, Basso G, Antonarakis SE, Jee R, Cotter F, Nizetic D (1998) Increased levels of a chromosome 21-encoded tumour invasion and metastasis factor (TIAM1) mRNA in bone marrow of Down syndrome children during the acute phase of AML (M7) Genes Chromosomes Cancer 23:61–66
Kent WJ (2002) BLAT<PIC 001><PIC 002>The BLAST-like alignment tool. Genome Res 12:656–664
Kouzarides T, Ziff E (1989) Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature 340:568–571
Landschulz WH, Johnson PF, McKnight SL (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764
Lee YT (1983) Primary carcinoma of the liver: diagnosis, prognosis, and management. J Surg Oncol 22:17–25
Luscher B, Larsson LG (1999) The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene 18:2955–2966
Munch C, Ludolph AC (2001) Pharmacological treatment of ALS. Neurol Neurochir Pol 35 [Suppl 1) 41–50
Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1998) Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res 5:31–39
Perez S, Vial E, van Dam H, Castellazzi M (2001) Transcription factor ATF3 partially transforms chick embryo fibroblast by promoting growth factor-independent proliferation. Oncogene 20:1135–1141
Petti S, Testa U, Migliaccio AR, Mavilio F, Marinucci M, Lazzaro D, Russo G, Mastroberardino G, Peschle C (1985) Embryonic hemopoiesis in human liver: morphologic aspects at sequential stages of ontogenic development. Prog Clin Biol Res 193:57–71
Saito T, Guan F, Papolos DF, Lau S, Klein M, Fann CS, Lachman HM (2001) Mutation analysis of SYNJ1: a possible candidate gene for chromosome 21q22-linked bipolar disorder. Mol Psychiatry 6:387–395
Schiaffonati L, Tiberio L (1997) Gene expression in liver after toxic injury: analysis of heat shock response and oxidative stress-inducible genes. Liver 17:183–191
Sellers JW, Struhl K (1989) Change fos oncoprotein to a jun-independent DNA binding protein with GCN4 dimerization specificity by swapping “leucine zipper”. Nature 341:74–76
Smith MJ, Charron-Prochownik DC, Prochownik EV (1990) The leucine zipper of c-Myc is required for full inhibition of erythroleukemia differentiation. Mol Cell Biol 10:5333–5339
Sun Z, Zhu Y, Stjernsward J, Hilleman M, Collins R, Zhen Y, Hsia CC, Lu J, Huang F, Ni Z, et al (1991) Design and compliance of HBV vaccination trial on newborns to prevent hepatocellular carcinoma and 5-year results of its pilot study. Cancer Detect Prev 15:313–318
Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, Cai T, Zhang X, Xiao HS, Qu J, Liu F, Huang QH, Cheng ZH, Li NG, Du JJ, Hu W, Shen KT, Lu G, Fu G, Zhong M, Xu SH, Gu WY, Huang W, Zhao XT, Hu GX, Gu JR, Chen Z, Han ZG (2001) Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci 98:15089–15094
Yeh FS, Yu MC, Mo CC, Luo S, Tong MJ, Henderson BE (1989) Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer Res 49:2506–2509
Yu Y, Zhang C, Zhou G, Wu S, Qu X, Wei H, Xing G, Dong C, Zhai Y, Wan J, Ouyang S, Li L, Zhang S, Zhou K, Zhang Y, Wu C, He F (2001) Gene expression profiling in human fetal liver and identification of tissue- and developmental-stage-specific genes through compiled expression profiles and efficient cloning of full-length cDNAs. Genome Res 11:1392–1403
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, Z., Yu, L., Wu, H. et al. Identification of a novel liver-specific expressed gene, TCP10L, encoding a human leucine zipper protein with transcription inhibition activity. J Hum Genet 48, 556–563 (2003). https://doi.org/10.1007/s10038-003-0075-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-003-0075-6