Abstract
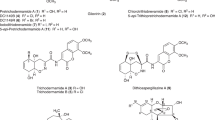
In our effort to find the key intermediates of lankacidin biosynthesis in Streptomyces rochei, three UV-active compounds were isolated from mutant FS18, a gene disruptant of lkcA encoding a non-ribosomal peptide synthetase (NRPS)-polyketide synthase (PKS) hybrid enzyme. Their structures were elucidated on the basis of spectroscopic data of NMR and MS. Two compounds of a higher mobile spot on silica gel TLC (Rf=0.45 in CHCl3-MeOH=20:1) were determined to be an epimeric mixture of citreodiol and epi-citreodiol at the C-6 position in the ratio of 2:1. In contrast, the compound of a lower mobile spot (Rf=~0 in CHCl3-MeOH=20:1) was identical to a 28-membered polyene macrolide pentamycin. The yields of citreodiols and pentamycin in FS18 were 5- and 250-fold higher compared with the parent strain. Introduction of a second mutation of srrX, coding a biosynthetic gene of the signaling molecules SRBs, into mutant FS18 did not affect the production of three metabolites. Thus, their production was not regulated by the SRB signaling molecules in contrast to lankacidin or lankamycin.
Similar content being viewed by others
Introduction
The filamentous soil bacteria Streptomyces species produce a vast array of secondary metabolites including antibacterial, antifungal, anticancer, antiparasitic, herbicidic and immunosuppressant agents. Recent development of the genome sequencing strategy revealed that Streptomyces strains usually contain over 20 secondary metabolite biosynthetic gene clusters, many of which are cryptic and silent under laboratory culture condition.1 Hence, Streptomyces genome is well considered to be an utmost important reservoir of natural products for new pharmaceutical applications.
Streptomyces rochei 7434AN4 produces two structurally unrelated polyketide antibiotics, lankacidin and lankamycin (Figure 1), and carries three linear plasmids, pSLA2-L, -M and -S.2 Lankacidin and lankamycin inhibit peptide synthesis synergistically by binding to the neighboring sites in the large ribosomal subunit,3, 4 and their biosyntheses are strictly controlled by the small diffusible signaling molecules termed SRBs (Streptomyces rochei butenolides) in S. rochei.5 In particular, lankacidin is a unique 17-membered carbocyclic polyketide with various biological activities.6 Nucleotide sequencing of the largest plasmid pSLA2-L (210 614 bp) together with extensive mutational analysis revealed that the lankacidin biosynthetic gene (lkc) cluster was located on pSLA2-L.7, 8, 9, 10 The lkc cluster contains a non-ribosomal peptide synthetase (NRPS)/polyketide synthase (PKS) hybrid gene (lkcA), three multidomain PKSs (lkcC, lkcF, lkcG), discrete dehydratase (lkcB) and acyltransferase (lkcD) genes, an amine oxidase gene (lkcE) and a gene cluster for pyrroloquinoline quinone (PQQ) biosynthesis (lkcK-lkcO). Recently, involvement of an additional dehydratase activity in lankacidin biosynthesis was reported by Dickschat et al.11
It is noteworthy that the lkc cluster contains only five ketosynthase domains, although eight rounds of condensation are required for the construction of the lankacidin skeleton, suggesting the modular-iterative mixed polyketide biosynthesis of lankacidin. We carried out extensive mutations on the lkc-PKS genes to isolate the key intermediates supporting this hypothesis.8, 10 Although we have not yet succeeded to isolate such intermediates, we noticed that the lkcA disruptant FS18 produced large amounts of UV-active compounds compared with the parent strain. We reported here the isolation and structural elucidation of citreodiol (5a), epi-citreodiol (5b) and pentamycin (6) from this mutant. In addition, we also describe that the production of the three metabolites was not regulated by the SRB signaling molecules based on the result of an additional mutation of srrX.
Results
Metabolites from the lkcA disruptant FS18
The lkcA mutant, termed FS18, exhibited complex metabolic changes in comparison with the parent and other lkc-PKS mutants. Strain FS18 failed to produce lankacidin and its derivatives due to the mutation in the early biosynthetic step of lankacidin (Figure 2). As for the lankamycin production, FS18 produced a similar level of lankamycin (8.2 mg l−1) compared with the parent strain 51252 (8.5 mg l−1). Remarkably, this strain accumulated the large amounts of UV-active compounds (5 and 6) (Figures 2a and c) and exhibited significant antifungal activity against Aspergillus niger (Figure 2d).
Analysis of the metabolites produced by strain FS18. (a) HPLC chromatograms acquired using photodiode array detector. (I) strain 51252 (parent), (II) strain FS18 (ΔlkcA). The crude extract was applied on a COSMOSIL 5C18-MS-II column (4.6 × 250 mm) and eluted with a mixture of acetonitrile-10 mM sodium phosphate buffer (pH 8.2) (3:7, v/v) at a flow rate of 1.0 ml min−1. (b) The UV-visible spectra of compounds 1-3, 5a, 5b and 6. (c) Silica gel TLC analysis of metabolites from 51252 (parent), FS18 (ΔlkcA) and KA78 (ΔlkcAΔsrrX) when irradiated with UV lamp at 254 nm (I) and stained with anisaldehyde (II). TLC plate was developed with a mixture of CHCl3-MeOH (15:1, v/v). (d) Bioassay of crude extracts against Aspergillus niger. The plate was incubated at 28 °C for 5 days.
The UV-active compound 5, a spot with strong UV-absorbance on TLC (Rf=0.45 in CHCl3-MeOH=20:1), was successively purified by Sephadex LH20 with MeOH and silica gel chromatography with CHCl3-MeOH=50:1-30:1. This compound was detected at 5.9 min on HPLC with a maximum absorbance at 267 nm, which differed from that of lankacidin antibiotics (λmax=230 nm) (Figures 2a and b). High-resolution positive ESI-MS gave a distinct molecular ion peak at 237.1096 [M+Na]+ (calcd. for C11H18O4Na, 237.1097). However, 22 carbon signals were observed in its 13C-NMR spectrum, which suggests that this compound is a mixture of inseparable isomers in the ratio of 2:1 (5a:5b). To separate 5a and 5b, a protection–deprotection strategy was carried out using carbonyldiimidazole in toluene according to Shizuri et al.12 as shown in Figure 3. The first step quantitatively yielded a mixture of carbonate derivatives 7a and 7b, which were then separated by silica gel chromatography (Rf=0.55 for 7a and 0.50 for 7b in hexane-EtOAc=2:1). Deprotection of the carbonate group by treatment with 0.5 M NaOH in THF-MeOH (1:1, v/v) afforded diastereomerically pure 5a and 5b. From their spectral data, compounds 5a and 5b were identical to citreodiol (5a; Figure 1) and epi-citreodiol (5b), both of which were originally isolated from Penicillium citreoviride B.12 As no complete set of NMR assignments has been reported, the 1H and 13C spectral data were summarized in Table 1. The optical rotation values of 5a and 5b were [α]D25=−4.8 (c=0.21, CHCl3) and [α]D26=+6.1 (c=0.16, CHCl3), respectively. All spectral data of 5a and 5b were identical to the reported values for synthetic (6S,7S)(+)-citreodiol and (6R,7S)(-)-epi-citreodiol, except for their opposite rotation values; [α]D22=+4.3 (c=1.0, CHCl3) for (+)-citreodiol and [α]D21=−7.1 (c=2.3, CHCl3) for (−)-epi-citreodiol.12, 13, 14 Thus, compounds 5a and 5b were elucidated to be (6R,7R,2E,4E)-6,7-dihydroxy-2,6-dimethylocta-2,4-dienoic acid methyl ester and (6S,7R,2E,4E)-6,7-dihydroxy-2,6-dimethylocta-2,4-dienoic acid methyl ester, respectively. This is the first discovery of 5a and 5b from Streptomyces species. Average yield of 5 in FS18 was calculated to be 1.2 mg l−1 from the HPLC peak intensity, which was fivefold larger compared with the parent strain 51252.
Compound 6 with strong UV absorbance was detected as a blue-gray spot on TLC when baked after anisaldehyde-H2SO4 spraying (Rfs=~0 in CHCl3-MeOH=20:1 and 0.44 in CHCl3-EtOAc-MeOH-H2O=22:22:44:11), and appeared as a distinct peak at 28.8 min on HPLC (Figures 2a and c). Its UV spectrum (λmax at 322, 338, 356 nm) (Figure 2b) was characteristic of methylpentaenes such as filipin and pentamycin.15, 16 This compound was successively purified by Sephadex LH20 with MeOH and a preparative HPLC with acetonitrile-10 mM sodium phosphate buffer (pH 8.2). Its molecular formula was determined by high-resolution positive ESI-MS to be C35H58O12 (observed m/z of 693.3810, calcd. for C35H58O12Na [M+Na]+=693.3821). In its 13C NMR, 35 carbons were classified into 3 methyl, 9 methylene, 21 methine and 2 quaternary carbons. Methine carbons were further classified into 9 pentaene carbons and 12 deshielded carbons. All spectral data (Supplementary Table S1) agreed well with the reported data of pentamycin (= fungichromin) (6, Figure 1).17, 18 Pentamycin is a 28-membered pentaene macrolide with a strong antifungal activity,16 suggesting that the observed inhibitory zone against A. niger in Figure 2d was caused by the action of 6. Average yield of 6 in FS18 was 5.0 mg l−1, while the relative yield in strain 51252 was estimated to be 0.02 mg l−1 from the calibration curve of 6. Hence, mutation in the early step of lankacidin biosynthesis showed 250-fold improvement in pentamycin production.
Synthesis of pentamycin, citreodiol and epi-citreodiol was not regulated by the SRB signaling molecules
In many Streptomyces species, antibiotic production is strictly controlled by the small diffusible signaling molecules such as A-factor for streptomycin production in Streptomyces griseus19 and virginia butanolides for virginiamycin production in Streptomyces virginiae.20 Two signaling molecules, SRB1 and SRB2, were isolated from S. rochei and their structures of SRB1 and SRB2 were determined to be (1′R)2-(1′-hydroxyl-6′-oxo-8′-methylnonyl)-3-methyl-4-hydroxybut-2-en-1,4-olide and (1′R)2-(1′-hydroxyl-6′-oxo-8′-methyldecyl)-3-methyl-4-hydroxybut-2-en-1,4-olide, respectively.5 SRB1 and SRB2 induced lankacidin and lankamycin production at 40 nM concentration.5 It is noteworthy that the biosynthetic gene clusters of lankacidin and lankamycin as well as their regulatory genes including srrX, which encodes a synthetic enzyme for SRBs, are located on pSLA2-L in S. rochei.
To analyze whether the synthesis of compounds 5a, 5b and 6 is regulated by the SRB signaling molecules, a second mutation of srrX was introduced into strain FS18. The BamHI site near the 5'-end of srrX was filled-in by Klenow enzyme (Figure 4a), which caused a frame shift in srrX. Plasmid pKAR3069 containing the mutated srrX gene on the Streptomyces-E. coli shuttle vector pRES1821 was transformed into strain FS18 to give a lkcA-srrX double mutant KA78. The frame-shift mutation on srrX was confirmed by Southern hybridization analysis. As shown in Figure 4b, the 5.6-kb BamHI and 1.9-kb BamHI-EcoRI fragments in strain FS18 were replaced by the 7.5-kb BamHI-EcoRI fragment in KA78. Metabolites produced by strain KA78 were analyzed by TLC, HPLC and bioassay against A. niger (Figures 2 and 4). This strain produced 5a, 5b and the antifungal agent 6 at a similar level to the single lkcA mutant FS18 (Figure 4c), while failed to accumulate lankacidins 1-3 or lankamycin 4 (Figure 2c). The single srrX mutant could not produce compounds 5a, 5b and 6 as well as 1-4. This result showed that production of three compounds (5a, 5b, 6) was not regulated by the SRB signaling molecules.
Construction of the lkcA-srrX double mutant KA78 and analysis of its metabolites. (a) Construction of the lkcA-srrX double mutant KA78. Ec, EcoRI; Ba, BamHI, Hi, HindIII; tsr, thiostrepton resistant gene. (b) Southern blot analysis of total DNA. Lane 1, λ/HindIII; lane 2, strain FS18/BamHI; lane 3, strain KA78/BamHI. (c) HPLC chromatograms of metabolites from strains 51252, FS18, KA78 and KY85 (the srrX single mutant) monitored at 267 nm (I) and at 338 nm (II). The crude extract was applied on a COSMOSIL 5C18-MS-II column (4.6 × 250 mm) and eluted with a mixture of acetonitrile-10 mM sodium phosphate buffer (pH 8.2) (3:7, v/v) at a flow rate of 1.0 ml min−1.
Discussion
In this study, we analyzed complex metabolic changes and the structures of citreodiol (5a), epi-citreodiol (5b) and pentamycin (6) overproduced by the lkcA disruptant FS18 of S. rochei. The LkcA protein plays an important role in both recognition of glycine as a starter unit and its condensation with a malonyl CoA extender unit in the early step of lankacidin biosynthesis.8 Altering the metabolic flux of fatty acid precursors sometimes causes an enhanced production of polyketides in the producing microorganisms.22 In the case of the polyether antibiotic nanchangmycin in Streptomyces nanchangensis, its titer was improved threefold by gene inactivation of the uncharacterized PKS clusters.23 For heterologous expression of secondary metabolite gene clusters, Komatsu et al.24 constructed versatile model host Streptomyces avermitilis SUKA17, whose chromosomal region including principal biosynthetic genes for avermectin and oligomycin was extensively deleted. Consequently, expression of the introduced oxytetracycline genes in the SUKA17 transformant was increased to yield a fivefold titer compared with the wild-type transformant. However, the overproduction of pentamycin and citreodiols in S. rochei seems different from these cases, because it occurred only in the lkcA mutant. None of the other genes in the lkc cluster, including three ketoreductase domains (lkcC-KR, lkcF-KR1 and lkcF-KR2), a C-methyltransferase gene (lkcC-MT), discrete acyltransferase (lkcD) and dehydratase (lkcB) genes, caused an overproduction when they were disrupted. These results suggest that the improved production of 5a, 5b and 6 in FS18 is not related to the cease of lankacidin production, though they are synthesized from a common precursor, malonyl CoA. The molecular mechanism of complex metabolic changes in FS18 remains to be clarified.
Citreodiol (5a) and epi-citreodiol (5b) were originally isolated from ascomycetous fungi Penicillium citreoviride B with a concomitant production of citreoviridin, a potent inhibitor of ATP synthesis and ATP hydrolysis.12 Some of the secondary metabolic pathways are common in bacteria and fungi; for example, cephalosporins (a member of β-lactam antibiotics) are produced by both actinomycetes (Streptomyces clavuligerus) and fungi (Acremonium chrysogenum).25 Biosynthetic genes for citreodiols have not been identified in P. citreoviride B or S. rochei. Comparative study will provide intriguing insights how secondary metabolite clusters have evolved among prokaryotic Streptomyces and eukaryotic fungi. Compounds 5a and 5b were produced even by the pSLA2-L-deficient strain UV1-2,2 harboring pSLA2-M and -S (data not shown). Sequence analysis revealed that pSLA2-M and -S have no structural genes for polyketide synthesis (accession numbers; AB597522 and AB905437, respectively).26 Thus, the biosynthetic genes for 5a and 5b may be located on the S. rochei chromosome.
Pentamycin (6) was originally isolated from Streptomyces cellulosae ATCC12625 (as fungichromin)27 and Streptomyces penticus (as pentamycin),16 and its absolute configuration was determined by spectral comparison of the degradation products and partial synthesis.28, 29 A trace amount of pentamycin was also isolated from a lankacidin-producing strain of Streptomyces rochei var. volubilis.30 Filipin III, a 14-deoxy congener of pentamycin, is the most studied pentaene antibiotic from biological31, 32 and biosynthetic33 aspects. Its biosynthetic gene cluster (pte) in Streptomyces avermitilis spans over 80 kb and contains 13 genes.34 Homologous genes with the pte cluster were not found on pSLA2-L, -M and -S, indicating the chromosomal location of the pentamycin gene (pem) cluster. Indeed, the pem cluster was identified in the contig sequences of strain 7434AN4, which contains 15 genes with an additional P450 monooxygenase gene pemI and a ferredoxin gene pemJ (data not shown). Further studies on the pem cluster are in progress in our laboratory.
Mutation of the srrX gene in strain FS18 revealed that the production of citreodiol, epi-citreodiol and pentamycin is not regulated by the SRB signaling molecules. In S. rochei, the signaling molecules/receptor system, SRBs/SrrA, strictly controls lankacidin and lankamycin production.35, 36, 37 All of the genes responsible for biosynthesis and regulation of the two antibiotics are coded on the linear plasmid pSLA2-L. Streptomyces coelicolor A3(2) contains two signaling molecule/receptor systems, SCB1/ScbR38 and methylenomycin furan (MMF)/MmfR.39 The former regulates the cryptic modular PKS gene cluster (kas; SCO6273-SCO6288) coded on the chromosome, while the latter does the methylenomycin biosynthetic genes coded on the linear plasmid SCP1. Hence, it is plausible that production of the three metabolites is controlled by an unidentified signaling molecule/receptor system coded on the S. rochei chromosome. Together with the synergistic action of lankacidin and lankamycin to the neighboring sites on ribosome, the location of their biosynthetic clusters and regulatory genes on the linear plasmid pSLA2-L show ingenious strategies of Streptomyces in genome evolution and environmental adaptation.
Materials and methods
Strains and culture conditions
S. rochei wild-type strain 7434AN4 and strain 51252 that carries only pSLA2-L were described previously.2, 7 Strain FS18, a disruptant of the NRPS/PKS hybrid gene lkcA, was constructed previously (Supplementary Figure S1).8 In strain FS18, the 1.6-kb kanamycin cassette was introduced into the BamHI site (nt 33 166 of pSLA2-L) on the lkcA gene (nt 31 013–35 881 complement of pSLA2-L). YEME liquid medium (0.3% yeast extract, 0.5% polypeptone, 0.3% malt extract, 1% D-glucose and 34% sucrose) was used for preparation of S. rochei protoplasts.40 Protoplasts were regenerated on R1M agar medium.41 YM medium (0.4% yeast extract, 1.0% malt extract and 0.4% D-glucose, pH 7.3) was used for antibiotic production. Tryptic soy broth (TSB) medium was used for preparation of Aspergillus niger spore suspension and bioassay. Escherichia coli XL1-Blue (Agilent Technologies, Santa Clara, CA, USA) was used for construction of targeting plasmids. E. coli strains were grown in Luria Bertani (LB) medium supplemented with ampicillin (100 μg ml−1) for plasmid maintenance when necessary.
Detection and isolation of metabolites
Detailed procedures were described in Supplementary Information.
Construction of the lkcA-srrX double mutant KA78
A 1.8-kb PCR fragment containing the srrX gene was amplified using cosmid C7 (nt 143 101–184 445 of pSLA2-L)7 as a template and a pair of primers, srrX-f1 (5′-GACGAATTCGTGAACGCGCCGGCCACCAG-3′, where italics indicate an EcoRI site) and srrX-r1 (5′-CTGAAGCTTGTAGCCGCGCTCGTTGCGCC-3′, where italics indicate a HindIII site). This fragment was digested with EcoRI and HindIII and cloned into pUC19 to give pKAR3067. The BamHI restriction site near the 5′-end of srrX was eliminated by filling-in and self-ligating reactions to give pKAR3068, the vector of which was replaced by E. coli-Streptomyces shuttle vector pRES1821 to give a targeting plasmid, pKAR3069.
This plasmid was transformed into protoplasts of S. rochei FS18. Thiostrepton-resistant transformants were subjected to sequential cultivation in YM liquid medium supplemented with thiostrepton (10 μg ml−1) to give the plasmid-integrated (single crossover) strain, which was then cultured in the absence of thiostrepton to give the double crossover strain. Mutation was confirmed by Southern hybridization using a DIG DNA Labeling and Detection Kit (Roche Diagnostics, Germany) according to the manufacturer’s protocol.
Antifungal activity
Aspergillus niger was used as test microorganism. A spore suspension of A. niger was obtained by washing a well-grown Petri dish culture (90ø × 8 mm) with a sterile aqueous solution containing 1% Tween 20 (0.5 ml). The resulting suspension was diluted with sterile water (10 ml) and filtered through a cotton plug. The bioassay plate was composed of two layers; the bottom layer contained TSB medium with 1.5% agar, while the top agar contained TSB-agar (0.8%) supplemented with 1% spore suspension of A. niger. Crude extracts from 100-ml broth (40 μl) were dropped onto paper disks (diameter; 0.8 cm), put on the bioassay plates and incubated at 28 °C for 5 days. The relative yield of 6 in strain 51252 was estimated by the calibration curve of inhibition zones obtained by purified 6.
References
Nett, M., Ikeda, H. & Moore, B. S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26, 1362–1384 (2009).
Kinashi, H., Mori, E., Hatani, A. & Nimi, O. Isolation and characterization of large linear plasmids from lankacidin-producing Streptomyces species. J. Antibiot. 47, 1447–1455 (1994).
Auerbach, T. et al. The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics. Proc. Natl Acad. Sci. USA 107, 1983–1988 (2010).
Belousoff, M. J. et al. Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit. Proc. Natl Acad. Sci. USA 108, 2717–2722 (2011).
Arakawa, K., Tsuda, N., Taniguchi, A. & Kinashi, H. The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. ChemBioChem. 13, 1447–1457 (2012).
Harada, S., Higashide, E., Fugono, T. & Kishi, T. Isolation and structures of T-2636 antibiotics. Tetrahedron Lett. 10, 2239–2244 (1969).
Mochizuki, S. et al. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48, 1501–1510 (2003).
Arakawa, K., Sugino, F., Kodama, K., Ishii, T. & Kinashi, H. Cyclization mechanism for the synthesis of macrocyclic antibiotic lankacidin in Streptomyces rochei. Chem. Biol. 12, 249–256 (2005).
Tatsuno, S., Arakawa, K. & Kinashi, H. Analysis of modular-iterative mixed biosynthesis of lankacidin by heterologous expression and gene fusion. J. Antibiot. 60, 700–708 (2007).
Tatsuno, S., Arakawa, K. & Kinashi, H. Extensive mutational analysis of modular-iterative mixed polyketide biosynthesis of lankacidin in Streptomyces rochei. Biosci. Biotechnol. Biochem. 73, 2712–2719 (2009).
Dickschat, J. S. et al. An additional dehydratase-like activity is required for lankacidin antibiotic biosynthesis. ChemBioChem 12, 2408–2412 (2011).
Shizuri, Y., Nishiyama, S., Imai, D. & Yamamura, S. Isolation and stereostructures of citreoviral, citreodiol, and epicitreodiol. Tetrahedron Lett. 25, 4771–4774 (1984).
Ghosh, P., Cusick, J. R., Inghrim, J. & Williams, L. J. Silyl-substituted spirodiepoxides: stereoselective formation and regioselective opening. Org. Lett. 11, 4672–4675 (2009).
Giovannini, P. P., Fantin, G., Massi, A., Venturi, E. & Pedrini, P. Enzymatic diastereo- and enantioselective synthesis of α-alkyl-α,β-dihydroxyketones. Org. Biomol. Chem. 9, 8038–8045 (2011).
Whitfield, G. B., Brock, T. D., Ammann, A., Gottlieb, D. & Carter, H. E. Filipin, an antifungal antibiotic: Isomation and properties. J. Am. Chem. Soc. 77, 4799–4801 (1955).
Umezawa, S., Tanaka, Y., Ooka, M. & Shiotsu, S. A new antifungal antibiotic, pentamycin. J. Antibiot. 11, 26–29 (1958).
Noguchi, H. et al. Biosynthesis and full NMR assignment of fungichromin, a polyene antibiotic from Streptomyces cellulose. J. Am. Chem. Soc. 110, 2938–2945 (1988).
Li, Z., Rawlings, B. J., Harrison, P. H. & Vederas, J. C. Production of new polyene antibiotics by Streptomyces cellulosae after addition of ethyl (Z-16-phenylhexadec-9-enoate. J. Antibiot. 42, 577–584 (1989).
Ohnishi, Y., Kameyama, S., Onaka, H. & Horinouchi, S. The A-factor regulatory cascade leading to streptomycin production in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34, 102–111 (1999).
Kawachi, R. et al. Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol. Microbiol. 36, 302–313 (2000).
Ishikawa, J., Niino, Y. & Hotta, K. Construction of pRES18 and pRES19, Streptomyces-Escherichia coli shuttle vectors carrying multiple cloning sites. FEMS Microbiol. Lett. 145, 113–116 (1996).
Olano, C., Lombó, F., Méndez, C. & Salas, J. A. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab. Eng. 10, 281–292 (2008).
Sun, Y. et al. 'Streptomyces nanchangensis', a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148, 361–371 (2002).
Komatsu, M. et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth. Biol. 19, 384–396 (2013).
Aharonowitz, Y., Cohen, G. & Martin, J. F. Penicillin and cephalosporin biosynthetic genes: Structure, organization, regulation, and evolution. Annu. Rev. Microbiol. 46, 461–495 (1992).
Yang, Y. et al. pSLA2-M of Streptomyces rochei is a composite linear plasmid characterized by self-defense genes and homology with pSLA2-L. Biosci. Biotechnol. Biochem. 75, 1147–1153 (2011).
Tytell, A. A., McCarthy, F. J., Fisher, W. P., Bolhofer, W. A. & Charney, J. Fungichromin and fungichromatin: new polyene antifungal agents. Antibiot. Annu. 716–718 (1954–1955).
Oishi, T. Studies directed towards the stereoselective synthesis of polyene macrolide antibiotics. Pure Appl. Chem. 61, 427–430 (1989).
Matsumoto, K., Shimagaki, M., Nakata, T. & Oishi, T. Synthesis of acyclic polyol derivatives via enzyme-mediated aldol reaction. Tetrahedron Lett. 34, 4935–4938 (1993).
Harada, S., Kishi, T. & Mizuno, K. Studies on T-2636 antibiotics. II. Isolation and chemical properties of T-2636 antibiotics. J. Antibiot. 24, 13–22 (1971).
Wachtler, V. & Balasubramanian, M. K. Yeast lipid rafts? -an emerging view. Trends Cell Biol. 16, 1–4 (2006).
Butler, J. D. et al. Niemann-pick variant disorders: comparison of errors of cellular cholesterol homeostasis in group D and group C fibroblasts. Proc. Natl Acad. Sci. USA 84, 556–560 (1987).
Xu, L. H. et al. Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis. J. Biol. Chem. 285, 16844–16853 (2010).
Ikeda, H., Shin-ya, K. & Omura, S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 41, 233–250 (2014).
Arakawa, K., Mochizuki, S., Yamada, K., Noma, T. & Kinashi, H. γ-Butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology 153, 1817–1827 (2007).
Yamamoto, S., He, Y., Arakawa, K. & Kinashi, H. γ-Butyrolactone-dependent expression of the SARP gene srrY plays a central role in the regulatory cascade leading to lankacidin and lankamycin production in Streptomyces rochei. J. Bacteriol. 190, 1308–1316 (2008).
Suzuki, T., Mochizuki, S., Yamamoto, S., Arakawa, K. & Kinashi, H. Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci. Biotechnol. Biochem. 74, 819–827 (2010).
Takano, E. et al. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56, 465–479 (2005).
O'Rourke, S. et al. Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol. Microbiol. 71, 763–778 (2009).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces genetics, The John Innes Foundation: Norwich, UK, (2000).
Zhang, H., Shinkawa, H., Ishikawa, J., Kinashi, H. & Nimi, O. Improvement of transformation system in Streptomyces using a modified regeneration medium. J. Ferment. Bioeng. 83, 217–221 (1997).
Acknowledgements
We thank Professor John C. Vederas (University of Alberta) for a kind gift of natural pentamycin. We thank Mrs. Tomoko Amimoto (Natural Science Center for Basic Research and Development, Hiroshima University) for the measurement of high-resolution mass spectra. This work was supported by a Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), and a Charitable Trust Araki Medical and Biochemical Research Memorial Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Cao, Z., Yoshida, R., Kinashi, H. et al. Blockage of the early step of lankacidin biosynthesis caused a large production of pentamycin, citreodiol and epi-citreodiol in Streptomyces rochei. J Antibiot 68, 328–333 (2015). https://doi.org/10.1038/ja.2014.160
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2014.160
This article is cited by
-
Characterization and engineering of Streptomyces griseofuscus DSM 40191 as a potential host for heterologous expression of biosynthetic gene clusters
Scientific Reports (2021)
-
Screening and Purification of Natural Products from Actinomycetes that Induce a “Rounded” Morphological Phenotype in Fission Yeast
Natural Products and Bioprospecting (2021)
-
The genome sequence of Streptomyces rochei 7434AN4, which carries a linear chromosome and three characteristic linear plasmids
Scientific Reports (2019)
-
Manipulation of metabolic pathways controlled by signaling molecules, inducers of antibiotic production, for genome mining in Streptomyces spp.
Antonie van Leeuwenhoek (2018)