Abstract
The growth and metastasis of prostate cancer are regulated by prostate stroma through the tumor–stromal cell interactions. Small molecules that modulate the tumor–stromal cell interactions will be new anticancer drugs. In the course of our screening of the modulators, we isolated two new atpenins, NBRI23477 A (4) and B (5), from the fermentation broth of Penicillium atramentosum PF1420. Compounds 4 and 5 as well as atpenin A4 (1), A5 (2) and B (3) inhibited the growth of human prostate cancer DU-145 cells in the coculture with human prostate stromal cells more strongly than that of DU-145 cells alone.
Similar content being viewed by others
Introduction
The growth and metastasis of prostate cancer are regulated by prostate stroma.1, 2 We have reported earlier that prostate stromal cell (PrSC) promotes the growth of human prostate cancer cells through the secretion of insulin-like growth factor-I.3, 4 There is a possibility that small molecules could inhibit cancer cell growth by modulating tumor–stromal cell interactions. We developed the in vitro coculture system of human prostate cancer cells and PrSC, in which the growth of prostate cancer cell is increased by the coculture with PrSC.3, 5 Using the assay method, we have been searching for the modulators of the tumor–stromal cell interactions. In the course of our screening of the modulators, we isolated new atpenins, NBRI23477 A (4) and B (5), along with the known compounds, atpenin A4 (1), A5 (2) and B (3).6, 7 Here we describe the isolation, structure determination and biological activity of 4 and 5. We also report the activity of 1, 2 and 3 on our assay.
Materials and methods
Reagents
Rhodanile blue was purchased from Aldrich (Milwaukee, WI, USA). Insulin and hydrocortisone were obtained from Sigma (St Louis, MO, USA). Transferrin was obtained from Wako Pure Chemical Industries (Tokyo, Japan). The recombinant human basic fibroblast growth factor was purchased from Pepro Tech (London, UK).
Cells
The human prostate cancer DU-145 cells were obtained from American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (ICN Biomedicals, Aurora, OH, USA), 100 U ml−1 penicillin G and 100 μg ml−1 streptomycin at 37 °C with 5% CO2. The human normal PrSCs were obtained from Bio Whittaker (Walkersville, MD, USA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U ml−1 penicillin G, 100 μg ml−1 streptomycin, ITH (5 μg ml−1 insulin, 5 μg ml−1 transferrin and 1.4 μM hydrocortisone) and 5 ng ml−1 human basic fibroblast growth factor at 37 °C with 5% CO2.
Coculture experiment
A microplate assay method for the selective measurement of epithelial tumor cells in coculture with stromal cells using rhodanile blue dye was performed as described before.5 PrSCs were first inoculated into 96-well plates at 5000 cells per well in 100 μl of Dulbecco's modified Eagle's medium supplemented with ITH and 0.1% fetal bovine serum in the presence of various concentrations of the test compounds. After 2 days, 10 μl of DU-145 cell suspension (5000 cells) in serum-free Dulbecco's modified Eagle's medium was inoculated onto a monolayer of PrSC, and the cells were further cultured for 3 days. For monoculture of DU-145 cells, the assay medium alone was first incubated in the presence of test compounds for 2 days at 37 °C. Then, DU-145 cells were inoculated as described above and cultured for further 3 days.
Analytical measurement
Melting points were obtained on a Yanagimoto micro melting point apparatus (Yanagimoto, Kyoto, Japan). Optical rotations were measured on a JASCO P-1030 polarimeter (JASCO, Tokyo, Japan). UV spectra were recorded on a Hitachi 228 A spectrometer (Hitachi, Tokyo, Japan). 1H- and 13C-NMR spectra were measured on a JEOL JNM A400 spectrometer (JEOL, Tokyo, Japan) using TMS as an internal standard. High resolution electrospray ionization mass spectrometry (HR-ESI-MS) spectra were measured with a JEOL JMS-T100LC spectrometer (JEOL).
Fermentation of fungal strain PF1420
Penicillium atramentosum PF1420 was isolated from a soil sample collected in Iwamizawa, Hokkaido, Japan. A slant culture of P. atramentosum PF1420 was used to inoculate 100-ml Erlenmeyer flasks. Each contained 20 ml of a seed medium consisting of 2.0% soluble starch, 1.0% glucose, 0.2% soybean meal, 0.6% wheat germ, 0.5% polypeptone, 0.3% yeast extract and 0.2% CaCO3 in deionized water adjusted to pH 7.2 with NaOH solution before sterilization. The flasks were incubated at 25 °C for 72 h on a rotary shaker at 220 r.p.m. Portions of 1.0 ml of this seed culture were transferred into six 500-ml Erlenmeyer flasks, each of which contained 100 ml of a seed medium. The flasks were incubated at 25 °C for 48 h on a rotary shaker at 220 r.p.m. Portions of 150 ml of this seed culture were transferred into four stainless vats, each of which contained 2.5% soybean meal and water-absorbed rice (4 kg) as solid production medium. The stainless vats were thoroughly stirred and then statically cultured at 25 °C for 14 days. After incubation, 16-kg portion of the obtained culture was extracted with 32 l of 67% aqueous acetone.
Results
Isolation procedure for atpenins
The 16-kg culture broth of P. atramentosum PF1420 was extracted with 32 l of 67% aqueous acetone. The filtrate of the extracts was concentrated in vacuo to remove acetone. The aqueous solution (5 l, pH 7) was applied on an HP-20 column. After washing the column with H2O and 50% MeOH, active ingredients were eluted with 100% MeOH. The eluate was concentrated in vacuo, dissolved in 600 ml H2O and then extracted with EtOAc. The organic layer was dried over Na2SO4 and concentrated in vacuo to afford 9.92 g of dried material. The materials were applied on a silica gel column (450 g, Wakogel C-200, 75–150 μm; Wako, Osaka, Japan) prepared with CHCl3, and eluted with CHCl3 and CHCl3-MeOH. The fractions eluted with CHCl3-MeOH (25:1) were concentrated in vacuo to give 4.13 g of viscous material. The viscous material was applied on gel filtration chromatography of Sephadex LH-20 (MeOH). The fractions containing atpenins were concentrated in vacuo to give 2.76 g of crude material. The crude material was purified by a reversed-phase HPLC column (Inertsil ODS-3, 20 × 250 mm, 6.0 ml min−1) with 50% CH3CN to afford crude atpenins, 46.4 mg of 1, 119.8 mg of 2, 111.1 mg of 3 and 4, and 14.5 mg of 5. The crude sample of 5 was applied on a Sephadex LH-20 column (MeOH) to afford pure 8.0 mg of 5. The crude mixture of 3 and 4 was further purified by a reversed-phase HPLC column (Inertsil ODS-3, 10 × 250 mm, 3.0 ml min−1) with 75% CH3CN and 0.1% TFA, and then the fractions containing 3 and 4 were applied on a Sephadex LH-20 column (MeOH) to afford pure 72.4 mg of 3 and 4.5 mg of 4, respectively. The crude mixture of 1 and 2 was further purified by a reversed-phase HPLC column (Inertsil ODS-3, 10 × 250 mm, 3.0 ml min−1) with 60% CH3CN and 0.1% TFA, and then the fractions containing 1 and 2 were applied on a Sephadex LH-20 column (MeOH) to afford pure 15.5 mg of 1 and 26.1 mg of 2, respectively. Separation by analytical HPLC (Inertsil ODS-3, 3 μm, 4.6 × 150 mm, 1.0 ml min−1) with 75% CH3CN and 0.1% TFA gave the following retention times (in minutes): 4.38 (1), 4.40 (2), 5.29 (3), 4.95 (4) and 4.28 (5).
Physicochemical properties
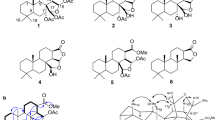
The physicochemical properties of 4 and 5 are summarized in Table 1. They are soluble in MeOH and DMSO. The molecular formulae of 4 and 5 were determined to be C15H21NO5Cl2 and C15H21NO5, respectively, by HR-ESI-MS. The general features of their UV and NMR spectra resembled each other, indicating structural similarities of these compounds. Compounds 1–3 were identified by NMR spectra as atpenin A4 (1), A5 (2) and B (3), respectively (Figure 1).6, 7
Structure determination of NBRI23477 A (4)
The 1H- and 13C-NMR data of 4 (Table 2) were similar to those of 2.8 However, the signals of 5′-methine and 6′-methylene of 2 were not observed in the 13C-NMR spectrum of 4, but a quaternary carbon (δ 97.9) and a methyl carbon (δ 35.7) appeared in 4. The partial structure of 4 was established by analyses of 1H-1H correlation spectroscopy (COSY) and heteronuclear multiple bond connectivity (HMBC) spectra (Figure 2). 1H-1H COSY spectrum revealed the following fragment: -CH(CH3)-CH2-CH(CH3)-. In the HMBC spectrum, singlet methyl protons (6′-H, δ 2.14) correlated to a quaternary carbon (C-5′, δ 97.9) and a methine carbon (C-4′, δ 48.0). Methyl protons (δ 1.35) connecting to C-2′ methine correlated to a carbonyl carbon (C-1′, δ 210.6). Therefore, the total structure of 4 was found to be a new family of atpenin (Figure 1).
Structure determination of NBRI23477 B (5)
The 13C- and 1H-NMR data of 5 (Table 2) were similar to those of 3.6, 7 However, the signals of 5′-methylene and 6′-methyl of 3 were not observed in the 13C-NMR spectrum of 5. On the other hand, an olefine carbon (δ 145.2) and a terminal olefine carbon (δ 114.4) were observed in 5. The partial structure of 5 was established by analyses of the 1H-1H COSY and HMBC spectra (Figure 3). 1H-1H COSY spectrum revealed the following fragment: -CH(CH3)-CH2-CH(CH3)-CH=CH2. In the HMBC spectrum, a methyl proton (δ 1.33) connecting to C-2′ methine correlated to a carbonyl carbon (C-1′, δ 211.3). Therefore, the total structure of 5 was found to be a new family of atpenin (Figure 1).
Biological activities
The effects of 1–5 on coculture of human prostate cancer DU-145 cells with PrSC were determined using rhodanile blue staining method.5 In the coculture, the growth of DU-145 cells is increased by PrSC.3, 5 As shown in Figure 4, all compounds showed selective growth inhibitory activities and inhibited the growth of DU-145 cells in coculture with PrSC more strongly than that of DU-145 cells alone. The IC50 values of 1–5 against the growth of DU-145 cells in coculture were 0.21, 0.021, 0.034, 0.064 and 0.13 μg ml−1, respectively, whereas those of 1–5 against the growth of DU-145 cells alone were 0.85, 0.048, 0.54, 0.95 and 0.71 μg ml−1, respectively. All compounds did not show apparent cytotoxicity against stromal cells under microscopic observation (data not shown).
Effects of 1–5 on coculture of DU-145 cells and PrSC. The growth of DU-145 cells cocultured with PrSC (•) or that of DU-145 cells alone (○) in the presence of the indicated concentrations of 1–5 was determined using rhodanile blue method. Values are means of duplicate determinations. Each s.e. is less than 10%.
Discussion
In this study, we have also obtained three structurally related compounds in addition to 1–5. The HR-ESI-MS spectra revealed that the molecular formulae of these compounds were C15H22NO6Cl, C15H23NO6 and C15H20NO5Cl3, respectively. Among them, a compound having the molecular formula of C15H20NO5Cl3 may be identical to reported WF-16775 A2,8 but we could not elucidate the structures of additional three compounds further due to their trace amounts. Ōmura et al.6 reported that there were atpenins A1, A2 and A3 along with A4, A5 and B, but they did not obtain A1, A2 and A3 in pure form and did not show any structural information. We cannot exclude the possibility that 4 and 5 would be identical to one of them. However, we have actually presented here two new structures of atpenins.
Atpenins A4, A5 and B were originally isolated as antifungal antibiotics.6, 7 Thereafter, atpenin B was found to decrease the cellular adenosine 5′-triphosphate.9 Furthermore, it is reported that atpenins specifically inhibit mitochondrial complex II (succinate–ubiquinone oxidoreductase).10 Mitochondria is now considered as a rational target for cancer therapy.11 Although there is a possibility that atpenins modulate tumor–stromal cell interactions by inhibiting mitochondrial functions, the elucidation of the precise mechanism of action needs to be studied further. We are now studying the effects of atpenins on tumor growth in vivo using mouse xenograft models.
References
Grossfeld, G. D., Hayward, S. W., Tlsty, T. D. & Cunha, G. R. The role of stroma in prostatic carcinogenesis. Endocr. Relat. Cancer 5, 253–270 (1998).
Tuxhorn, J. A., Ayala, G. E. & Rowley, D. R. Reactive stroma in prostate cancer progression. J. Urol. 166, 2472–2483 (2001).
Kawada, M., Inoue, H., Masuda, T. & Ikeda, D. Insulin-like growth factor I secreted from prostate stromal cells mediates tumor–stromal cell interactions of prostate cancer. Cancer Res. 66, 4419–4425 (2006).
Kawada, M., Inoue, H., Arakawa, M. & Ikeda, D. Transforming growth factor-β1 modulates tumor–stromal cell interactions of prostate cancer through insulin-like growth factor-I. Anticancer Res. 28, 721–730 (2008).
Kawada, M. et al. A microplate assay for selective measurement of growth of epithelial tumor cells in direct coculture with stromal cells. Anticancer Res. 24, 1561–1568 (2004).
Ōmura, S. et al. Atpenins, new antifungal antibiotics produced by Penicillium sp. Production, isolation, physico-chemical and biological properties. J. Antibiot. 41, 1769–1773 (1988).
Kumagai, H., Nishida, H., Imamura, N., Tomoda, H. & Ōmura, S. Structures of atpenins A4, A5 and B, new antifungal antibiotics produced by Penicillium sp. J. Antibiot. 43, 1553–1558 (1990).
Otsuka, T., Takase, S., Terano, H. & Okuhara, M. New angiogenesis inhibitors, WF-16755A1 and A2. J. Antibiot. 45, 1970–1973 (1992).
Oshino, K., Kumagai, H., Tomoda, H. & Ōmura, S. Mechanism of action of atpenin B on Raji cells. J. Antibiot. 43, 1064–1068 (1990).
Miyadera, H. et al. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate–ubiquinone oxidoreductase). Proc. Natl Acad. Sci. USA 100, 473–477 (2003).
Kim, J. & Dang, C. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66, 8927–8930 (2006).
Acknowledgements
We thank Dr R Sawa (Microbial Chemistry Research Center) for analysis of HR-ESI-MS spectra, and Ms K Adachi and Ms E Satoh for their technical assistance. This work was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kawada, M., Momose, I., Someno, T. et al. New atpenins, NBRI23477 A and B, inhibit the growth of human prostate cancer cells. J Antibiot 62, 243–246 (2009). https://doi.org/10.1038/ja.2009.20
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2009.20
Keywords
This article is cited by
-
The therapeutic potential of mitochondrial toxins
The Journal of Antibiotics (2021)
-
Novel approaches for identification of anti-tumor drugs and new bioactive compounds
The Journal of Antibiotics (2018)
-
Small molecules modulating tumor–stromal cell interactions: new candidates for anti-tumor drugs
The Journal of Antibiotics (2016)
-
Intervenolin, a new antitumor compound with anti-Helicobacter pylori activity, from Nocardia sp. ML96-86F2
The Journal of Antibiotics (2013)
-
NBRI16716A, a new antitumor compound against human prostate cancer cells, produced by Perisporiopsis melioloides Mer-f16716
The Journal of Antibiotics (2010)