Abstract
Ammocidins B, C and D were isolated from the culture broth of Saccharothrix sp. AJ9571, an ammocidin A-producing strain. Their structures were determined by a detailed spectroscopic analysis and by a comparison of their NMR data with those of ammocidin A. Ammocidins A and B showed potent anti-proliferative activities against human cancer cell lines.
Similar content being viewed by others
Introduction
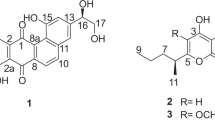
In the course of our screening for apoptosis inducers in Ras-dependent Ba/F3-V12 cells, we isolated ammocidin A (formerly named ammocidin) from the culture broth of Saccharothrix sp. AJ9571.1, 2 Ammocidin A induced apoptotic cell death in Ras-dependent Ba/F3-V12 cells with an IC50 of 66 ng ml−1, and no cell death was observed in IL-3-dependent Ba/F3-V12 cells at less than 100 μg ml−1 of ammocidin A. Further searching for congeners from the ammocidin-producing strain resulted in the isolation of ammocidins B, C and D (Figure 1). In this paper, we report the isolation, structure elucidation and cytotoxic activities of the ammocidins.
Results
Production and isolation
The seed medium was composed of yeast extract 0.1%, beef extract 0.1%, N-Z amine type A 0.2% and glucose 1.0% (pH 7.3). Saccharothrix sp. AJ9571 was cultured in flasks containing the seed medium on a rotary shaker at 30 °C for 4 days. The resultant seed culture at 2.0% was transferred into 500-ml Erlenmeyer flasks containing 100 ml of a production medium consisting of glycerol 2.0%, starch 1.0%, rape seed meal 2.0%, soytone 0.3% and calcium carbonate 0.3% (pH 7.0). The fermentation was carried out on a rotary shaker at 30 °C for 7 days. The culture filtrate (10 l) was extracted with ethyl acetate and the extract was subjected to silica gel column chromatography with chloroform–methanol (8 : 1). The active eluate was chromatographed on a Sephadex LH-20 column with methanol. The active fraction was purified by HPLC using a Senshu Pak PEGASIL ODS column with 70% methanol. Further purification was carried out on the same column with 33% acetonitrile to give ammocidins A (387 mg), B (24 mg), C (15 mg) and D (24 mg).
Physico-chemical properties
The physico-chemical properties of ammocidins B, C and D are summarized in Table 1. The molecular formulae of ammocidins B, C and D were determined by high-resolution electrospray ionization mass spectra to be C60H98O22, C53H86O18 and C46H74O16, respectively. The IR spectra indicated the presence of hydroxyl (3430 cm−1) and conjugated carbonyl groups (1670–1720 cm−1). The UV spectra of ammocidins B, C and D are characterized by a maximum absorption around 320 nm.
Structure elucidation
The structures of ammocidins B, C and D were established by 1H- and 13C-NMR and two-dimensional NMR experiments (Correlation Spectroscopy (COSY), Heteronuclear multiple quantum coherence (HMQC), heteronuclear multiple-bond correlation (HMBC)) on a 600-MHz spectrometer. The 1H and 13C-NMR assignments of ammocidins B, C and D in CD3OD are listed in Table 2.
The 1H-NMR spectrum of ammocidin B showed the presence of 12 methyl groups between 1.10 and 2.08 p.p.m. and four methoxy groups between 3.24 and 3.53 p.p.m. The 13C-NMR spectrum confirmed the presence of 60 carbons. 1H spin networks from 7-H to 9-H, from 15-H to 20-H, from 22-H to 23-H and from 25-H to 28-H, and 1H–13C long-range correlations as shown in Figure 2 revealed the aglycone structure of ammocidin B, which is the same as ammocidin A. Moreover, a 1H spin network from an anomeric proton (1′-H) to 6′-H and an heteronuclear multiple-bond correlation between 1′-H and C-5′ revealed the presence of 6-deoxyglucose, which is the same component as that of ammocidin A. Two 2,6-dideoxy-3-methylpyranose moieties were constructed by 1H spin networks from an anomeric proton 1″-H (1′″-H) to 2″-H (2′″-H) and from H-4″ (H-4′″) to 6″-CH3 (6′″-CH3), and heteronuclear multiple-bond correlation from 3″-CH3 (3′″-CH3) to C-2″ (C-2′″), C-3″ (C-3′″) and C-4″ (C-4′″). Vicinal proton coupling constants (Table 2) and an NOE between 1″-H (1′″-H) and 3″-CH3 (3′″-CH3) identified these sugars as β-olivomycose. The three glycoside linkages were formed on the basis of long-range couplings between 1′-H and C-9, between 1″-H and C-24, and between 1′″-H and C-4″. These data indicate that ammocidin B is an ammocidin A derivative containing the second β-olivomycose residue in place of the β-digitoxose residue (Figure 1).
The 1H- and 13C-NMR spectra of ammocidin C were similar to those of ammocidin B, although signals due to the terminal olivomycose residue (C-1′″–C-6′″) were absent in ammocidin C. In the aglycone, signals indicative of a methylene group (δC 32.2, δH 2.58, 1.98) appeared in place of those for the oxymethine (C-16) in ammocidin B. A 1H spin network from 15-H to 20-H identified ammocidin C as a 16-deoxyaglycone metabolite. The molecular formula and the upfield shift of C-4″ (δC 90.3 in ammocidin B and δC 80.1 in ammocidin C) indicated that ammocidin C is the 16-deoxy-4″-O-deolivomycosyl derivative of ammocidin B. Finally, two-dimensional NMR analyses established the planar structure of ammocidin C as shown in Figure 1.
The 1H- and 13C-NMR spectra indicated that ammocidin D consists of the same aglycone as ammocidin B and 6-deoxyglucose as a sole sugar moiety. The molecular formula of ammocidin D and the upfield shift of 24-C (δC 82.2 in ammocidin B and δC 75.0 in ammocidin D) showed the loss of two olivomycose residues from ammocidin B. The planar structure of ammocidin D was confirmed as shown in Figure 1 by two-dimensional NMR data.
Cytotoxic activities of ammocidins
To determine the cytotoxic activities of the ammocidins, A549 human lung carcinoma cells, MCF-7 human breast carcinoma cells and HCT116 human colon carcinoma cells were tested. The cell proliferation assay showed potent cytotoxic activities of ammocidins A and B (Table 3). The IC50’s of ammocidins A and B were 0.06–0.4 μM.
Discussion
During our efforts to obtain congeners from the ammocidin-producing strain, we isolated new ammocidin derivatives, ammocidins B, C and D. The main structural differences of these compounds are the number of olivomycose residues attached to 24-O. Whereas ammocidin A contains a digitoxose residue and an olivomycose residue, the sugar chain of ammocidin B consisted of two olivomycose residues. The ammocidins are considered to be good tools to investigate the importance of sugar moieties for the biological activity of 20-membered macrolides.
We evaluated the anti-proliferative effects of ammocidins on human cancer cell lines. Although all ammocidins showed cytotoxic activities, ammocidins A and B were more potent than ammocidins C and D. Thus, deoxysugar moieties attached to 24-O play an important role in the anti-proliferative activities of ammocidins. Human cancer cell lines A549 and HCT116 harbor oncogenic ras mutation (A549: K-ras-G12S, HCT116: K-ras-G13D). MCF-7 cells, however, have no ras mutation (but harbor PI3KCA mutation). Ammocidins showed potent anti-proliferative activities against all the three cell lines, implying that their anti-proliferative activities might not be due to Ras inhibition. Further biological studies on ammocidins are in progress.
Methods
Cell culture
The cancer cell lines A549 (lung adenocarcinoma), MCF-7 (breast adenocarcinoma) and HCT-116 (colon carcinoma) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2 atmosphere.
Assay of anti-proliferative activity
Anti-proliferative activities were measured by using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega Corporation, Madison, WI, USA), which determines the number of viable cells in a culture based on quantitation of ATP. In brief, cancer cells were seeded at 1 × 103 cells per well in 96-well microplates and cultured overnight. The cells were treated with various concentrations of ammocidins for 48 h. After 50 μl per well of CellTiter-Glo Reagent was added to the cell culture, the luminescence was measured by a ARVO.sx multilabel counter (Wallac Berthold, Turku, Finland). IC50 values were determined from the dose–response curves of growth inhibition.
General experimental procedures
NMR spectra were obtained on a Varian NMR System 600 NB CL (Varian NMR Systems, Palo Alto, CA, USA). Methanol-d4 was used as a solvent and an internal reference. Melting points were determined with a Yanagimoto (Seisakusyo, Japan) micro melting point apparatus. High-resolution electrospray ionization mass spectra were recorded on a Waters LCT-Premier XE mass spectrometer (Waters, Milford, MA, USA). Optical rotations were recorded on a HORIBA SEPA-300 polarimeter (Horiba, Tokyo, Japan). UV spectra were obtained on a HITACHI U-3200 spectrophotometer (Hitachi, Tokyo, Japan). IR spectra were obtained using a HORIBA FT-720 spectrophotometer.
References
Murakami, R, Tomikawa, T, Shin-ya, K, Seto, H & Hayakawa, Y Ammocidin, a new apoptosis inducer in Ras-dependent cells from Saccharothrix sp. I. Production, isolation and biological activity. J Antibiot 54, 710–713 (2001).
Murakami, R, Tomikawa, T, Shin-ya, K, Seto, H & Hayakawa, Y Ammocidin, a new apoptosis inducer in Ras-dependent cells from Saccharothrix sp. II. Physico-chemical properties and structure elucidation. J Antibiot 54, 714–717 (2001).
Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan, and by a grant from the New Energy and Industrial Technology Development Organization (NEDO), Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Murakami, R., Shinozaki, J., Kajiura, T. et al. Ammocidins B, C and D, new cytotoxic 20-membered macrolides from Saccharothrix sp. AJ9571. J Antibiot 62, 123–127 (2009). https://doi.org/10.1038/ja.2008.23
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2008.23
Keywords
This article is cited by
-
Amycolatopsins A–C: antimycobacterial glycosylated polyketide macrolides from the Australian soil Amycolatopsis sp. MST-108494
The Journal of Antibiotics (2017)
-
Precursor-directed in situ synthesis of Saccharothriolides G and H by the Actinomycete Saccharothrix sp. A1506
The Journal of Antibiotics (2017)
-
Saccharothrix sp. PAL54, a new chloramphenicol-producing strain isolated from a Saharan soil
World Journal of Microbiology and Biotechnology (2012)