Abstract
Oral lichen planus (OLP) is a chronic inflammatory disease that is frequently detected in oral tissues. The aim of our study was to identify the prevalence of the detection of periodontopathogenic microorganisms (Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythia and Treponema denticola in OLP patients and to compare with this prevalence of periodontopathogenic microorganisms in healthy non-OLP patients. Our study included 27 (18 chronic periodontitis (OLPP) and 9 gingivitis (OLPG)) patients diagnosed with OLP along with 26 (13 chronic periodontitis (HP) and 13 gingivitis (HG)) healthy non-OLP patients. The multiplex polymerase chain reaction (PCR) with subsequent reverse hybridization method (micro-IDent) was used for identifying periodontopathogenic microorganisms present in subgingival plaque samples. The percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in subgingival plaque samples taken from OLP patients (OLPG and OLPP) were 18.5%, 85.1%, 81.4%, 88.8% and 74%, respectively. Meanwhile, in the non-OLP patients (HG and HP), these values were 7.6%, 50%, 46.1%, 73% and 57.7%, respectively. Thus, comparing the non-OLP groups with the OLP groups, the periodontopathogens’ percentages of detection in the OLP groups were higher than those in the non-OLP groups. According to our study results, OLP patients have higher levels of infection with A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola than non-OLP patients. We argue that the high percentages in patients with OLP may help identify the importance of periodontopathogenic microorganisms in the progress of periodontal diseases of OLP.
Similar content being viewed by others
Introduction
The oral mucosa is one region in which many types of dermatosis are frequently observed. These types of dermatosis mainly include lichen planus (LP), bullous pemphigoid, erythema multiforme and lupus erythematosus. LP is a chronic, inflammatory, and mucocutaneous disease that affects 0.9%–1.2% of the population.1,2 LP affects mucous membranes, especially the oral mucosa and/or skin. Oral lichen planus (OLP), a oral form of LP, is found in reticular, papular, plaque-like, atrophic and ulcerative forms;3,4 it is observable in children but it is frequently a disease of middle age, mainly observed in women compared to men,4 and it demonstrates malignant transformations at rates of 0.3%–10%.5,6 Approximately 70% of OLP oral lesions are observed in the buccal mucosa, palatinal mucosa, tongue, lip mucosa, alveolar mucosa and gingiva.7,8,9 The etiological factors of OLP are mental stress, malnutrition, infection (viral), mechanical trauma and tobacco use.10,11 Psychological evaluation should be conducted to identify the etiology of the disease.12,13 Various medicines and chemicals may cause OLP or OLP-like reactions11 Virusesthat play a role in the etiology of OLP include the herpes viruses, especially the Epstein–Barr Virus,14 human papillomavirus,15 and hepatitis C virus (HCV).16,17 Although it has been determined that OLP and viruses are related, the roles of bacteria in the etiology of OLP have not yet been determined. However, it is known that OLP is related to periodontal diseases.
The culture method has been used to detect bacterial species in subgingival plaque. However, the culture method is inefficient in the identification of anaerobic microorganisms.18 Recently, various methods have been used in the assay of microorganisms. Molecular methods are used in the identification of microbial composition in subgingival plaque samples. One such method is multiplex polymerase chain reaction (PCR) followed by DNA–DNA hybridization. Periodontopathogenic microorganisms (Aggregatibactor actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythia, Treponema denticola) have been identified with a micro-IDent kit (Hain Lifescience GmbH, Nehren, Germany) multiplex PCR with subsequent reverse hybridization method.19
It is known that patients with OLP are more likely to be infected by many other microorganisms.20 However, the role of periodontopathogenic microorganisms (A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia, T. denticola) in the pathogenesis of OLP has not been clearly identified.
The purpose of this study was to use the micro-IDent kit to compare periodontopathogenic microorganisms (A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia, T. denticola) in 27 OLP (18 OLP periodontitis (OLPP) and 9 OLP gingivits (OLPG)) patients with 26 healthy non-OLP (13 chronic periodontitis (HP) and 13 gingivitis (HG)) patients.
Materials and methods
Study population
A total of 27 OLP patients who fit the criteria of our study were randomly chosen as the study group from patients who were referred to the clinic at the Department of Periodontology, Faculty of Dentistry, Selcuk University. They were histologically and clinically diagnosed with OLP at the Başkent University Konya Practice and Research Hospital’s Department of Dermatology clinic. The patients within the non-OLP group that took part in our study were randomly selected from those that were found to be suitable according to the criteria of our study; all had reported for periodontal treatment in 2005–2006 to the clinic at the Selcuk University Faculty of Dentistry Department of Periodontology. This study was approved by the Human Ethics Research Committee of Selcuk University Faculty of Dentistry (2005-51). The patients were selected according to the clinical and radiographic criteria proposed by the 1999 International Workshop for the Classification of Periodontal Diseases and Conditions.21 Each subject read and signed an informed consent form and read the Helsinki Declaration before entering the study.
The OLP groups (test groups) consisted of 27 patients with OLP, and their diagnosis was histologically validated. In the test group of OLP patients, 18 patients were diagnosed with chronic periodontitis, and 9 were diagnosed with gingivitis. Form and localization were defined by evaluating the clinical lesions of OLP patients. Meanwhile, the dermatologically healthy non-OLP groups (control groups) consisted of 13 patients diagnosed with chronic periodontitis, and 13 with gingivitis, for a total of 26 systemically healthy non-OLP patients.
The criteria for patient selection were: no systematic disease, no antibiotics taken for at least 6 months before sampling, non-smokers and not pregnant. In the clinical evaluation, a probing depth of 4–6 mm and 6 mm or greater of clinical attachment loss in at least six regions was considered to indicate chronic periodontitis. Bleeding on probing in at least 50% of the total gingiva, no vertical or horizontal bone loss upon radiographic examinations (bone crest at >95% of proximal tooth sites and <3 mm between the cemento-enamel junction) were considered to indicate the presence of gingivitis. To evaluate the periodontal conditions of patients before phase-1 periodontal treatment, records for Plaque Index,22 Gingival Index,23 probing depth and clinical attachment loss were observed. All of the clinical measurements were conducted by a single researcher.
Sampling of subgingival plaque
After noting that no supragingival plaque deposits were present in sample sites, the sites were cleaned with sterile cotton pellets and isolated with sterile cotton rolls. The subgingival plaque samples were taken from two Ramfjord sample teeth (teeth 1.6, 2.1, 2.4, 3.6, 4.1 and 4.4) that had the deepest periodontal pocket formation in chronic periodontitis (OLPP and HP) and gingivitis (OLPG and HG) samples. The plaque samples weretransferred to Eppendorf tubes, each containing 140 µL buffer (10 mmol⋅L−1 Tris-HCl, 1.0 mmol·L−1 ethylene diaminetetraacetic acid (EDTA), pH 7.6).
Microbiological evaluation
Experiments were conducted at Division of Molecular Microbiology, Department of Microbiology and Clinic Microbiology, Selcuk University’s Meram Medical Faculty. An extraction kit (Invisorb, RTP Spin Bacteria DNA Mini Kit; Invitek, Berlin, Germany) was used to isolate bacterial DNA from plaque samples. DNA isolation was conducted according to the protocol suggested by the manufacturer. The presence of A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in DNA-isolated plaque samples was investigated by the micro-IDent microorganism kit. The total volume was prepared as a 50 µL mixture, according to the manufacturer’s recommendations. For the 50 µL mixture, a master mixture was prepared with 35 µL reaction mixture, 5 µL 10× PCR buffer, 5 µL MgCl2 and 0.4 µL DNA polymerase, with 5 µL of DNA sample added. The process for PCR reaction was as follows: following a 4-min holding time at 95 °C, 2 min for 10 cycles at 58 °C for one cycle at 95 °C, and a total of 20 cycles at 95 °C, 40 s at 53 °C and 40 s at 70 °C; the desired products were amplified with Gene Amp 9700 PCR (Applied Biosystems, Foster City, CA, USA) at an extension of 8 minutes at 70 °C. The presence of bacteria was determined on the PCR product with biotin, and nitrocellulose strips with the reverse hybridization method, according to the manufacturer’s suggestions, with ‘line probes’ specific to A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola. Strips that were developed by probes specific to bacteria were evaluated by colorimetric monitoring.
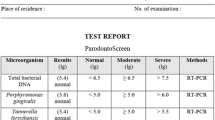
All obtained strips were scanned in a computer medium. Scanned strips were transferred to the Adobe Photoshop (Adobe Systems, San Jose, CA, USA) program and bands related to the microorganisms on the strips were analyzed. All of the contrast (autocontrast) luminescence amounts of the bands were measured. A white background represented 0% and the conjugate control on the strip was evaluated as 100%. The values obtained were separated into three groups: clear-looking band (0.01%–29.99%), weak-looking bands (30.00%–59.99%) and the invasible bands (50%–100.00%). The weak-looking band and the invisible band were considered to be positive for microorganisms. The clear-looking band was accepted as being negative, and the microorganism percentages were determined.
Statistical analyses
Data analyses were conducted using a statistical software package program (SPSS 14.0). All results were evaluated using a one-sample Kolmogorov–Smirnov test to identify the distribution. A Mann–Whitney U test was used to identify the differences in clinical periodontal parameters between groups. The significance value was defined as P<0.05.
Results
The ages, genders and periodontal clinical parameters of the groups are shown in Table 1. No significant difference was observed between groups (HG to OLPG and HP to OLPP) with regard to age and gender (P>0.05). Comparing both groups, a statistically significant difference was observed in the periodontal clinical parameters between the gingivitis groups (OLPG and HG) and the chronic periodontitis groups (OLPP and HP) (P<0.05). The chronic periodontitis groups (OLPP and HP) had higher periodontal clinical parameters compared to the gingivitis groups (OLPG and HG) (P<0.05). The OLPG patients had higher periodontal clinical parameters than the HG patients, but statistically significant difference was not observed (P>0.05). Similarly the OLPP patients, had a higher periodontal clinical parameters than the HP patients, but statistically significant difference was not observed (P>0.05).
At clinical examination, 9 patients with OLP had erosive-type lesions, 15 had reticular-type lesions and 3 had plaque type lesions.
The percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in the study groups are given in Figure 1. The percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in subgingival plaque samples taken from OLP patients (OLPG and OLPP) were 18.5%, 85.1%, 81.4%, 88.8% and 74%, respectively, and there values in the non-OLP groups (HG and HP) were 7.6%, 50%, 46.1%, 73% and 57.7%, respectively. The periodontopathogen microorganism’s percentages of detection in the OLP groups (OLPG and OLPP) were higher than those in the non-OLP groups (HG and HP).
Semiquantitative load of periodontopathogens in the healthy non-OLP gingivitis, OLP gingivitis, healthy non-OLP periodontitis and OLP periodontitis groups. Semiquantitative load determined by percentage of relative of densitometry controls, Low: 0.01%–9.99%; moderate: 10.00%–39.99%; high: 40%–69.99%; very high: ≥70.00%. Themultiplex PCR with subsequent reverse hybridization method (micro-IDent) was used in identifying A. actinomycetemcomitans, P. gingivalis and P. intermedia present in subgingival plaque samples. OLP, oral lichen planus; HG, healthy non-OLP gingivitis; HP, healthy non-OLP periodontitis; OLPG, OLP gingivitis; OLPP, OLP periodontitis; PCR, polymerase chain reaction.
If groups were considered separately, the percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola microorganisms in the HG group were defined as 0%, 23%, 15.3%, 69.2% and 23%, respectively. In the OLPG group, the percentage of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola were 0%, 66.6%, 55.5%, 66.6% and 22.2%, respectively. Percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in HP patients were defined as 15.3%, 77%, 77%, 92.3% and 77%, respectively, whereas in the OLPP group, the percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola were 27.7%, 94.4%, 94.4%, 100% and 100%, respectively. Comparing the non-OLP groups with the OLP groups, the percentages of microorganisms in the OLP groups were higher than those in the non-OLP groups. This reveals that OLP patients have a greater tendency to be infected with periodontal pathogens (A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola).
Discussion
It is known that dental plaque plays an important role in the etiology of periodontal diseases.24,25 Recently, there have been many investigations in the literature that addressed whether different systemic diseases are risk factors for periodontal diseases, and inversely, whether periodontal disease is a risk factor for systemic diseases.26,27,28,29 There are limited investigations in the literature on the relationship and/or interaction between dermatosis (a systemic diseases) and periodontal diseases.303132,33,34 OLP is one of the most frequently observed types of dermatosis in the oral mucosa. OLP lesions can persist for a long time and in some cases can turn into oral carcinoma.5,6 Due to these characteristics, studies on OLP significantly contribute to the understanding of oral pathology and periodontology. In our study, subgingival dental plaque was sampled from all patients, and in dental plaque samples Aa, Pg, Pi, Tf and Td were evaluated by the micro-IDent microorganism kit. The results indicated that patients with OLP were infected with periodontopathogenic microorganisms at higher percentages compared to patients in the non-OLP control groups. There are no studies in the literature that have examined the periodontal clinical assessments of OLP and non-OLP patients and that have compared periodontopathogenic microorganisms with the micro-IDent microorganism kit.
Several studies14,15,16,17 have investigated the relationships between different species of viruses and OLP. However, very few, if any, studies have examined the relationship between bacteria and OLP. When examining the relationships between OLP and viruses HCV was observed in biopsy samples taken from the lesion areas of OLP patients, which demonstrated either the direct effect of the virus or the immune response developed against the viral antigens.32,33 The biopsy samples taken from the lesion of HCV-positive OLP patients additionally showed that the HCV-specific T lymphocytes were concentrated in this area.34,35 In spite of this, some studies have suggested that instead of being dependent on the immune response developed against the viral antigens inside the lesion or on the virus itself, lesions are caused by the evolution of the long-term stimuli that are established in the immune system as a result of chronic inflammation, potentially producing pathological results.36,37 Some studies have determined that HCV-dependent OLP cases exhibited immunohistochemical differences fromOLP cases that were not dependent on HCV (refs. 38–39). The Epstein–Barr virus was observed to be present in 26% of tissue samples taken from OLP subjects, which was a significantly higher percentage than observed in controls.40 In another study that examined the relationship between human papillomavirus and OLP, it was determined that HPV was present in significantly higher levels in the OLP group relative to the control group.41 Herpes simplex virus-1 has occasionally been found in OLP, mainly in erosive lesions.42,43 As is evident from these studies, viruses are closely related with OLP lesions.
The current body of literature lacks studies that have investigated the relationship between A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia, T. denticola and OLP (ref. 20). Only a few studies have examined bacteria in OLP patients with molecular methods. Bornstein et al.20 evaluated the bacterial colonization in mucosa samples taken from OLP and non-OLP patients using DNA–DNA hybridization. In the samples taken from OLP lesion sites, significantly higher bacterial counts of Actinomyces odontolyticus, Campylobacter gracilis, Eikenella corrodens, Fusobacterium nucleatum naviforme, Fusobacterium nucleatum subsp. polymorphum, Lactobacillus acidophilus, Neisseria mucosa, Prevotella nigrescens, Selenomonas noxia, Staphylococcus aureus, Streptococcus gordonii, Streptococcus mitis, Prevotella intermedia, Streptococcus sanguinis, Tannerella forsythia, Veillonella parvula, Bacteroides ureolyticus, Dialister species, Staphylococcus haemolyticus, Streptococcus agalactiae, Bifidobacterium biavatii, Haemophilus influenzae and Lactobacillus crispatus were present when compared with samples from healthy controls.
However, several studies have examined periodontopathogenic microorganisms in different periodontal diseases using the micro-IDent microorganism kit.18,19 For example, Eick and Pfister19 compared periodontopathogenic microorganisms (A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia, T. denticola) using the micro-IDent kit and the cultivation method in plaque samples taken from patients with aggressive periodontitis and periodontally healthy patients. According to the their results, the percentage of detection of A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in periodontally healthy patients were 12%, 12%, 8%, 8% and 2%, respectively, whereas these detection rates in patients with aggressive periodontitis were 20%, 33%, 52%, 68% and 50%, respectively. In our study the percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in HP patients were 15.3%, 77%, 77%, 92.3% and 77%, respectively. When the percentages of detection were compared between these two studies, the A. actinomycetemcomitans observation percentage was observed to be higher, and the P. gingivalis, P. intermedia, T. forsythia and T. denticola observation percentages were found to be lower in our study. The reason for these differences is believed to be the fact that different microorganisms play a role in the pathogenesis of chronic periodontitis and aggressive periodontitis. Urban et al.18 compared the micro-IDent test kits with a conventional culture procedure, and according to the results, the micro-IDent test more often detected Tannerella forsythia compared to the conventional method. The micro-IDent kit detected almost the same number of samples positive for A. actinomycetemcomitans, P. gingivalis, P. intermedia and T. denticola as did the culture procedures. Derdilopoulou et al.44 compared periodontopathogenic microorganisms with the micro-IDent kit in subgingival plaque samples taken at 3-month intervals (before periodontal treatment, the third month after periodontal treatment and sixth month after periodontal treatment). In their study, periodontopathogens (A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia, T. denticola) were detected at 31.6% of the sites before periodontal treatment; these levels decreased after 3 months (26.0%), and increased again 6 months after therapy (32.6%). Eick et al.45 compared the micro-IDent test with real-time PCR methods in dental plaque samples taken at 3, 6 and 12 months post-therapy for patients with chronic periodontitis. According to the results, semiquantitative DNA-strip technology (micro-IDent) is more suitable for microbial analysis in chronic periodontitis patients. Socransky et al.46 compared microflora in periodontal pockets and they found a higher prevalence of P. gingivalis, P. intermedia, Pacifastacus nigrescens and T. denticola in deep periodontal pockets than in shallow periodontal pockets. Griffen et al.47 investigated the microorganism profile in healthy patients and patients with periodontal disease, and they found that P. gingivalis was detected in only 25% (46 of 181) of the healthy subjects, but was detected in 79% (103 of 130) of the periodontal disease group. In our study, the percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola microorganisms in patients in the HG group were 0%, 23%, 15.3%, 69.2% and 23%, respectively. In another study in which the percentages of microorganisms were investigated, the percentage of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in patients with chronic periodontitis were 22.2%, 96.3%, 63%, 96.3% and 100%, respectively; the percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in patients with aggressive periodontitis were 50%, 78.1%, 62.5%, 87.5% and 87.5%, respectively; and the percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in periodontally healthy control patients were 8.8%, 11.8%, 26.5%, 55.9% and 67.6%, respectively.48 In our study, the percentages of detection for A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola in HP patients were 15.3%, 77%, 77%, 92.3% and 77%, respectively. These percentages were similar between our study and that of Reichert et al.48.
Studies that investigated the periodontal status of patients with OLP indicated that local factors, such as dental plaque and dental calculus, impact the improvement of dermatological lesions in oral tissues, and lesions may have erosive forms in regions where local irritants are found in higher quantities.30,31,49 In our study, patients in the OLP group presented higher levels of periodontopathogenic microorganisms; specifically, OLPP patients exhibited higher percentages compared to patients in the HP group. The oral lesions in patients with OLP may lead to bleeding and may be painful. Due to the symptoms of OLP lesions, patients cannot adequatelyperform their oral hygiene habits. Thus, periodontal diseases progress more severely in OLP patients. The severity of periodontal disease increases with increased OLP lesion symptoms, and this in turn may lead to an increase in the amount of microorganisms (A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia and T. denticola) that are periodontopathogenic.
Conclusions
The amounts of periodontopathogens in OLP patients were found to be higher in comparison to non-OLP patients. Similar to viruses, periodontopathogenic microorganisms can also play a role in the etiology of OLP. The relationship between OLP and periodontopathogenic microorganisms can be explained by two hypotheses. The first is the increase in the numbers of periodontopathogens following the formation of OLP lesions; in other words, local factors such as dental plaques (which is the primary factor for periodontal disease) and dental calculus are local irritant factors that prevent the healing of OLP lesions, which alter the characteristics of the lesions into more aggressive forms. The numbers of periodontopathogenic microorganisms can increase depending on this situation. The second relationship is that periodontopathogeniv microorganisms play a direct role in the etiology of OLP and could cause the formation of OLP lesions. To understand the relationship between the OLP and periodontopathogenic microorganisms, further long-term clinical studies should be conducted.
References
Huber MA . Oral lichen planus. Quintessence Int 2004; 35( 9): 731–752.
Scully C, el-Kom M . Lichen planus: review and update on pathogenesis. J Oral Pathol 1985; 14( 6): 431–458.
Anuradha C, Reddy BV, Nandan SR et al. Oral lichen planus. A review. NY State Dent J 2008; 74( 4): 66–68.
Zakrzewska JM . Re: Mollaoglu N. Oral lichen planus: a review. Br J Oral Maxillofac Surg 2000; 38:370–377. Br J Oral Maxillofac Surg 2001; 39( 5): 407.
Holmstrup P, Thorn JJ, Rindum J et al. Malignant development of lichen planus-affected oral mucosa. J Oral Pathol 1988; 17( 5): 219–225.
Thorn JJ, Holmstrup P, Rindum J et al. Course of various clinical forms of oral lichen planus. A prospective follow-up study of 611 patients. J Oral Pathol 1988; 17( 5): 213–218.
Axell T, Rundquist L . Oral lichen planus—a demographic study. Community Dent Oral Epidemiol 1987; 15( 1): 52–56.
Bagan-Sebastian JV, Milian-Masanet MA, Penarrocha-Diago M et al. A clinical study of 205 patients with oral lichen planus. J Oral Maxillofac Surg 1992; 50( 2): 116–118.
Bhaskar SN . Synopsis of Oral Pathology. St Louis: The CV Mosby Company, 1986: 373–381.
Carrozzo M . How common is oral lichen planus? Evid Based Dent 2008; 9( 4): 112–113.
Gorsky M, Raviv M . Efficacy of etretinate (Tigason) in symptomatic oral lichen planus. Oral Surg Oral Med Oral Pathol 1992; 73( 1): 52–55.
Lundqvist EN, Wahlin YB, Bergdahl M et al. Psychological health in patients with genital and oral erosive lichen planus. J Eur Acad Dermatol Venereol 2006; 20( 6): 661–666.
Oliveira Alves MG, Almeida JD, Balducci I et al. Oral lichen planus: a retrospective study of 110 Brazilian patients. BMC Res Notes 2010; 3: 157.
Sand LP, Jalouli J, Larsson PA et al. Prevalence of Epstein–Barr virus in oral squamous cell carcinoma, oral lichen planus, and normal oral mucosa. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2002; 93( 5): 586–592.
Jontell M, Watts S, Wallstrom M et al. Human papilloma virus in erosive oral lichen planus. J Oral Pathol Med 1990; 19( 6): 273–277.
Konidena A, Pavani BV . Hepatitis C virus infection in patients with oral lichen planus. Niger J Clin Pract 2011; 14( 2): 228–231.
Zhou Y, Jiang L, Liu J et al. The prevalence of hepatitis C virus infection in oral lichen planus in an ethnic Chinese cohort of 232 patients. Int J Oral Sci 2010; 2( 2): 90–97.
Urban E, Terhes G, Radnai M et al. Detection of periodontopathogenic bacteria in pregnant women by traditional anaerobic culture method and by a commercial molecular genetic method. Anaerobe 2010; 16( 3): 283–288.
Eick S, Pfister W . Comparison of microbial cultivation and a commercial PCR based method for detection of periodontopathogenic species in subgingival plaque samples. J Clin Periodontol 2002; 29( 7): 638–644.
Bornstein MM, Hakimi B, Persson GR . Microbiological findings in subjects with asymptomatic oral lichen planus: a cross-sectional comparative study. J Periodontol 2008; 79( 12): 2347–2355.
Armitage GC . Development of a classification system for periodontal diseases and conditions. Ann Periodontol 1999; 4( 1): 1–6.
Silness J, Loe H . Periodontal disease in pregnancy. II. Correlation between oral hygiene and periodontal condition. Acta Odontol Scand 1964; 22: 121–135.
Löe H . The gingival index, the plaque index and the retention index systems. J Periodontol 1967; 38( 6): 610–616.
Haffajee AD, Cugini MA, Dibart S et al. Clinical and microbiological features of subjects with adult periodontitis who responded poorly to scaling and root planing. J Clin Periodontol 1997; 24( 10): 767–776.
Offenbacher S . Periodontal diseases: pathogenesis. Ann Periodontol 1996; 1( 1): 821–878.
Kiran M, Arpak N, Unsal E et al. The effect of improved periodontal health on metabolic control in type 2 diabetes mellitus. J Clin Periodontol 2005; 32( 3): 266–272.
Mustapha IZ, Debrey S, Oladubu M et al. Markers of systemic bacterial exposure in periodontal disease and cardiovascular disease risk: a systematic review and meta-analysis. J Periodontol 2007; 78( 12): 2289–2302.
Wilder RS, Iacopino AM, Feldman CA et al. Periodontal-systemic disease education in U.S. and Canadian dental schools. J Dent Educ 2009; 73( 1): 38–52.
Erpenstein H . Periodontal and prosthetic treatment in patients with oral lichen planus. J Clin Periodontol 1985; 12( 2): 104–112. .
Holmstrup P, Schiotz AW, Westergaard J . Effect of dental plaque control on gingival lichen planus. Oral Surg Oral Med Oral Pathol 1990; 69( 5): 585–590.
Ramon-Fluixa C, Bagan-Sebastian J, Milian-Masanet M et al. Periodontal status in patients with oral lichen planus: a study of 90 cases. Oral Dis 1999; 5( 4): 303–306.
Carrozzo M, Francia P, Gandolfo S et al. Increased frequency of HLA-DR6 allele in Italian patients with hepatitis C virus-associated oral lichen planus. Br J Dermatol 2001; 144( 4): 803–808.
Nagao Y, Kameyama T, Sata M . Hepatitis C virus RNA detection in oral lichen planus tissue. Am J Gastroenterol 1998; 93( 5): 850.
Pilli M, Penna A, Zerbini A et al. Oral lichen planus pathogenesis: a role for the HCV-specific cellular immune response. Hepatology 2002; 36( 6): 1446–1452.
Mega H, Jiang WW, Takagi M . Immunohistochemical study of oral lichen planus associated with hepatitis C virus infection, oral lichenoid contact sensitivity reaction and idiopathic oral lichen planus. Oral Dis 2001; 7( 5): 296–305.
Nagao Y, Sata M, Itoh K et al. Quantitative analysis of HCV RNA and genotype in patients with chronic hepatitis C accompanied by oral lichen planus. Eur J Clin Invest 1996; 26( 6): 495–498.
Lodi G, Carrozzo M, Hallett R et al. HCV genotypes in Italian patients with HCV-related oral lichen planus. J Oral Pathol Med 1997; 26( 8): 381–384.
Kirby AC, Lodi GL, Olsen I et al. Immunohistochemical and serological comparison of idiopathic and hepatitis C virus-associated forms of oral lichen planus. Eur J Oral Sci 1998; 106( 4): 853–862.
Al Robaee AA, Al Zolibani AA . Oral lichen planus and hepatitis C virus: is there real association? Acta Dermatovenerol Alp Panonica Adriat 2006; 15( 1): 14–19.
Sand LP, Jalouli J, Larsson PA et al. Prevalence of Epstein–Barr virus in oral squamous cell carcinoma, oral lichen planus, and normal oral mucosa. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2002; 93( 5): 586–592.
Yildirim B, Sengüven B, Demir C . Prevalence of herpes simplex, Epstein Barr and human papilloma viruses in oral lichen planus. Med Oral Patol Oral Cir Bucal 2011; 16( 2): 170–174.
Lodi G, Scully C, Carrozzo M et al. Current controversies in oral lichen planus: report of an international consensus meeting. Part 1. Viral infections and etiopathogenesis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2005; 100( 1): 40–51.
Cox M, Maitland N, Scully C . Human herpes simplex-1 and papillomavirus type 16 homologous DNA sequences in normal, potentially malignant and malignant oral mucosa. Eur J Cancer B Oral Oncol 1993; 29( B): 215–219.
Derdilopoulou FV, Nonhoff J, Neumann K et al. Microbiological findings after periodontal therapy using curettes, Er:YAG laser, sonic, and ultrasonic scalers. J Clin Periodontol 2007; 34( 7): 588–598.
Eick S, Straube A, Guentsch A et al. Comparison of real-time polymerase chain reaction and DNA-strip technology in microbiological evaluation of periodontitis treatment. Diagn Microbiol Infect Dis 2011; 69( 1): 12–20.
Socransky SS, Haffajee AD, Cugini MA et al. Microbial complexes in subgingival plaque. J Clin Periodontol 1998; 25( 2): 134–144.
Griffen AL, Becker MR, Lyons SR et al. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol 1998; 36( 11): 3239–3242.
Reichert S, Machulla HK, Klapproth J et al. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res 2008; 43( 1): 40–47.
van der Waal I . Non-plaque related periodontal lesions. An overview of some common and uncommon lesions. J Clin Periodontol 1991; 18( 6): 436–440.
Acknowledgements
This study has been performed with project support from the Turkey Scientific and Technological Research Council (project no. 106S340) and Selcuk University Coordination of Scientific Research (project no. 06202034). Publication of this manuscript is supported by Open Fund of State Key Laboratory of Oral Diseases, Sichuan University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Seckin Ertugrul, A., Arslan, U., Dursun, R. et al. Periodontopathogen profile of healthy and oral lichen planus patients with gingivitis or periodontitis. Int J Oral Sci 5, 92–97 (2013). https://doi.org/10.1038/ijos.2013.30
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijos.2013.30
Keywords
This article is cited by
-
Does oral lichen planus aggravate the state of periodontal disease? A systematic review and meta-analysis
Clinical Oral Investigations (2022)
-
Salivary MRP-8/14 and the presence of periodontitis-associated bacteria in children with bonded maxillary expansion treatment
Clinical Oral Investigations (2021)
-
Oral lichen planus: a microbiologist point of view
International Microbiology (2021)
-
Dysbiosis of saliva microbiome in patients with oral lichen planus
BMC Microbiology (2020)
-
Alteration of Streptococcus salivarius in Buccal Mucosa of Oral Lichen Planus and Controlled Clinical Trial in OLP Treatment
Probiotics and Antimicrobial Proteins (2020)